publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2026
-
 Short-term hormonal modulation with mifepristone does not induce oncogenic changes in the endometrium of BRCA1/2 pathogenic variant carriersMartin Widschwendter, Chiara Herzog, Mohammed Fatih Rasul, and 6 more authorsCommunications Medicine, 2026
Short-term hormonal modulation with mifepristone does not induce oncogenic changes in the endometrium of BRCA1/2 pathogenic variant carriersMartin Widschwendter, Chiara Herzog, Mohammed Fatih Rasul, and 6 more authorsCommunications Medicine, 2026Progesterone receptor antagonists such as mifepristone have emerged as candidates for breast cancer prevention, particularly in high-risk populations such as BRCA1/2 pathogenic variant carriers. However, their impact on endometrial safety remains insufficiently characterized, raising concerns about unopposed oestrogen stimulation in the setting of impaired DNA repair. This study reports secondary outcomes evaluating short-term endometrial effects of mifepristone in this high-risk population. We previously conducted a randomized, double-blind, placebo-controlled trial (NCT01898312) involving 45 premenopausal women with BRCA1/2 pathogenic variants. Participants received mifepristone (50 mg every other day, n = 30) or a non-hormonal comparator (n = 15) for three months. Here we present secondary outcomes from the trial: Paired endometrial biopsies from a subset of 14 participants were analysed using transcriptomic, DNA methylation, and cell-type deconvolution methods. Statistical comparisons were performed using paired and unpaired Wilcoxon tests. Here we show that mifepristone induces amenorrhea in all treated participants without increasing epithelial cell proportions, the compartment most relevant to endometrial carcinogenesis. Multi-omics analyses reveal no molecular signatures consistent with oncogenic transformation. DNA methylation and gene expression indices associated with endometrial cancer remain stable after treatment, even after adjusting for age and cell composition. Short-term mifepristone exposure does not produce molecular changes linked to endometrial carcinogenesis in BRCA1/2 pathogenic variant carriers. These findings provide important safety data for the future development of progesterone receptor modulators in cancer prevention. Long-term studies are needed to confirm these observations.
-
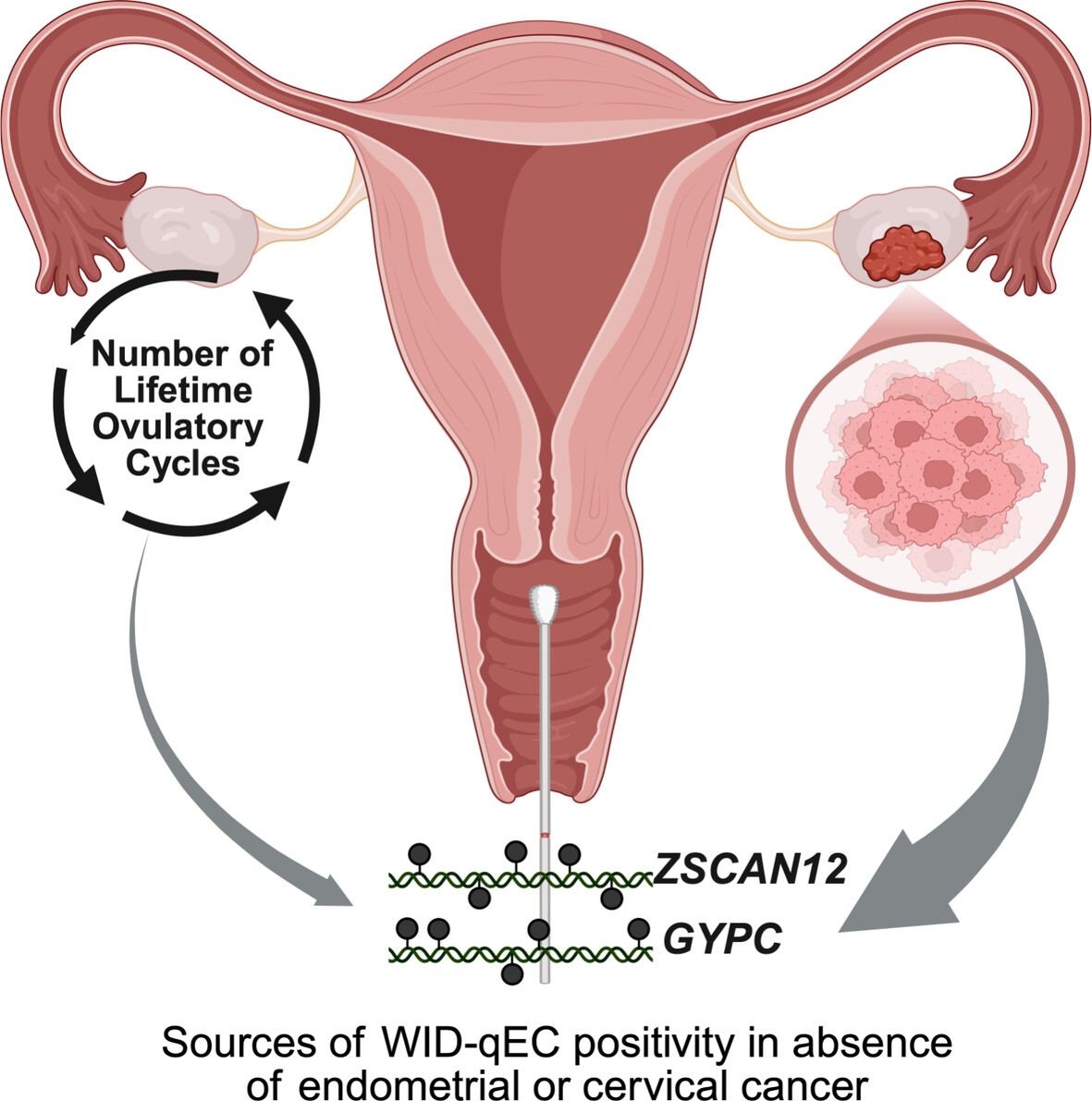 The cervico‐vaginal DNA methylation WID‐qEC test: An epigenetic marker associated with ovarian cancer in the absence of endometrial and cervical cancerElisa Redl, Chiara Herzog, Charlotte Vavourakis, and 16 more authorsInternational Journal of Cancer, 2026
The cervico‐vaginal DNA methylation WID‐qEC test: An epigenetic marker associated with ovarian cancer in the absence of endometrial and cervical cancerElisa Redl, Chiara Herzog, Charlotte Vavourakis, and 16 more authorsInternational Journal of Cancer, 2026The DNA methylation‐based WID‐qEC test, applied to cervico‐vaginal samples, has been validated for the accurate detection of endometrial and cervical cancers. However, a small proportion of women test positive despite the absence of these cancers. The aim of this study was to explore the biological and clinical characteristics associated with such WID‐qEC‐positive cases to inform potential follow‐up strategies. We analyzed 1269 cervico‐vaginal samples from women without endometrial or cervical cancer, including healthy controls (n = 624), women with benign gynecological conditions (n = 324), and ovarian cancer cases (n = 321). Of the 80 WID‐qEC‐positive results, 43 (54%) were from women with ovarian cancer. WID‐qEC positivity was associated with the presence of ovarian cancer (adjusted odds ratio [OR] 2.93; 95% CI 1.75–4.95) and with a higher number of lifetime ovulatory cycles (adjusted OR 2.67; 95% CI 1.06–7.50), a known ovarian cancer risk factor. Both associations were independent of age, menopausal status, hormone replacement therapy usage, or family history of breast or ovarian cancer. Our findings suggest that in the absence of endometrial or cervical cancer, WID‐qEC positivity may indicate an elevated risk or presence of ovarian cancer. While the standalone positive predictive value (PPV) for ovarian cancer detection remains low in the general population, we outline how WID‐qEC could be used in a two‐step triage approach. In women presenting with abnormal bleeding, combining WID‐qEC positivity with a highly specific plasma‐based cell‐free DNA methylation test (e.g., with 60%–80% sensitivity and \textbackslashtextasciitilde98.4% specificity) could theoretically yield a PPV of around 30%–40%. This hypothetical modeling is intended solely to illustrate how WID‐qEC positivity might inform future triage research, rather than to propose a clinical diagnostic algorithm. The DNA methylation‐based WID‐qEC test has been validated for the accurate detection of endometrial and cervical cancers using cervico‐vaginal samples. However, a small proportion of women test positive despite the absence of these cancers. This study explored the associated biological and clinical characteristics to inform potential follow‐up strategies. The results showed that in the absence of endometrial or cervical cancer, WID‐qEC test positivity may indicate an elevated risk or the presence of ovarian cancer. While the standalone positive predictive value for ovarian cancer detection remains low in the general population, WID‐qEC could potentially be used in future two‐step triage approaches.
-
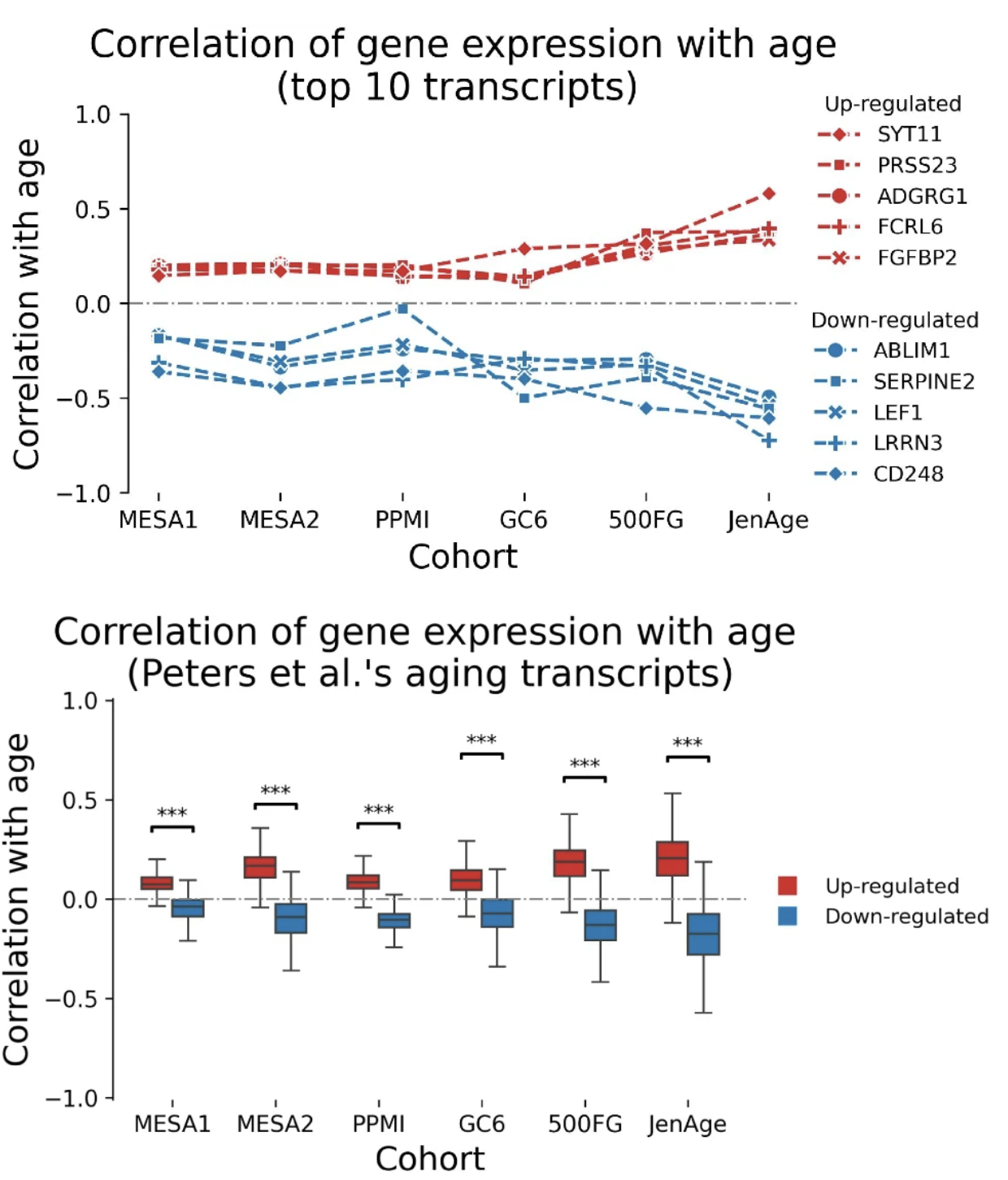 Integrative epigenetics and transcriptomics identify aging genes in human bloodMahdi Moqri, Kejun Ying, Jesse R. Poganik, and 16 more authorsNature Communications, 2026
Integrative epigenetics and transcriptomics identify aging genes in human bloodMahdi Moqri, Kejun Ying, Jesse R. Poganik, and 16 more authorsNature Communications, 2026Recent epigenome-wide studies have identified a large number of genomic regions that consistently exhibit changes in their methylation status with aging across diverse populations, but the functional consequences of these changes are largely unknown. On the other hand, transcriptomic changes are more easily interpreted than epigenetic alterations, but previously identified age-related gene expression changes have shown limited replicability across populations. Here, we develop an approach that leverages high-resolution multi-omic data for an integrative analysis of epigenetic and transcriptomic age-related changes and identify genomic regions associated with both epigenetic and transcriptomic age-dependent changes in blood. Our results show that these multi-omic aging genes in blood are enriched for adaptive immune functions, replicate more robustly across diverse populations and are more strongly associated with aging-related outcomes compared to the genes identified using epigenetic or transcriptomic data alone. These multi-omic aging genes may serve as targets for epigenetic editing to facilitate cellular rejuvenation.
2025
-
 Recommendations for biomarker data collection in clinical trials by longevity biotechnology companiesChiara M. S. Herzog, Jesse R. Poganik, Nicola Boekstein, and 6 more authorsnpj Aging, Dec 2025
Recommendations for biomarker data collection in clinical trials by longevity biotechnology companiesChiara M. S. Herzog, Jesse R. Poganik, Nicola Boekstein, and 6 more authorsnpj Aging, Dec 2025Biomarkers of aging have the potential to transform geroscience clinical trials because of their broad applications in stratifying participants, prioritizing interventions, and monitoring responses to geroprotectors. As longevity biotechnology companies (LBCs) continue to plan and launch innovative clinical trials, standard practices in collecting data and applying biomarkers of aging will allow the field to support parallel and ongoing validation and benchmarking efforts for aging biomarkers. Moreover, defining best practices will ensure future reuse of valuable clinical data through pre-competitive alignment on shared tools. Here, we propose recommendations for such collections. We believe that wide adoption of these recommendations will allow LBCs to produce and leverage the highest quality data from their clinical trials, while also benefiting the geroscience field more broadly with minimal additional effort.
-
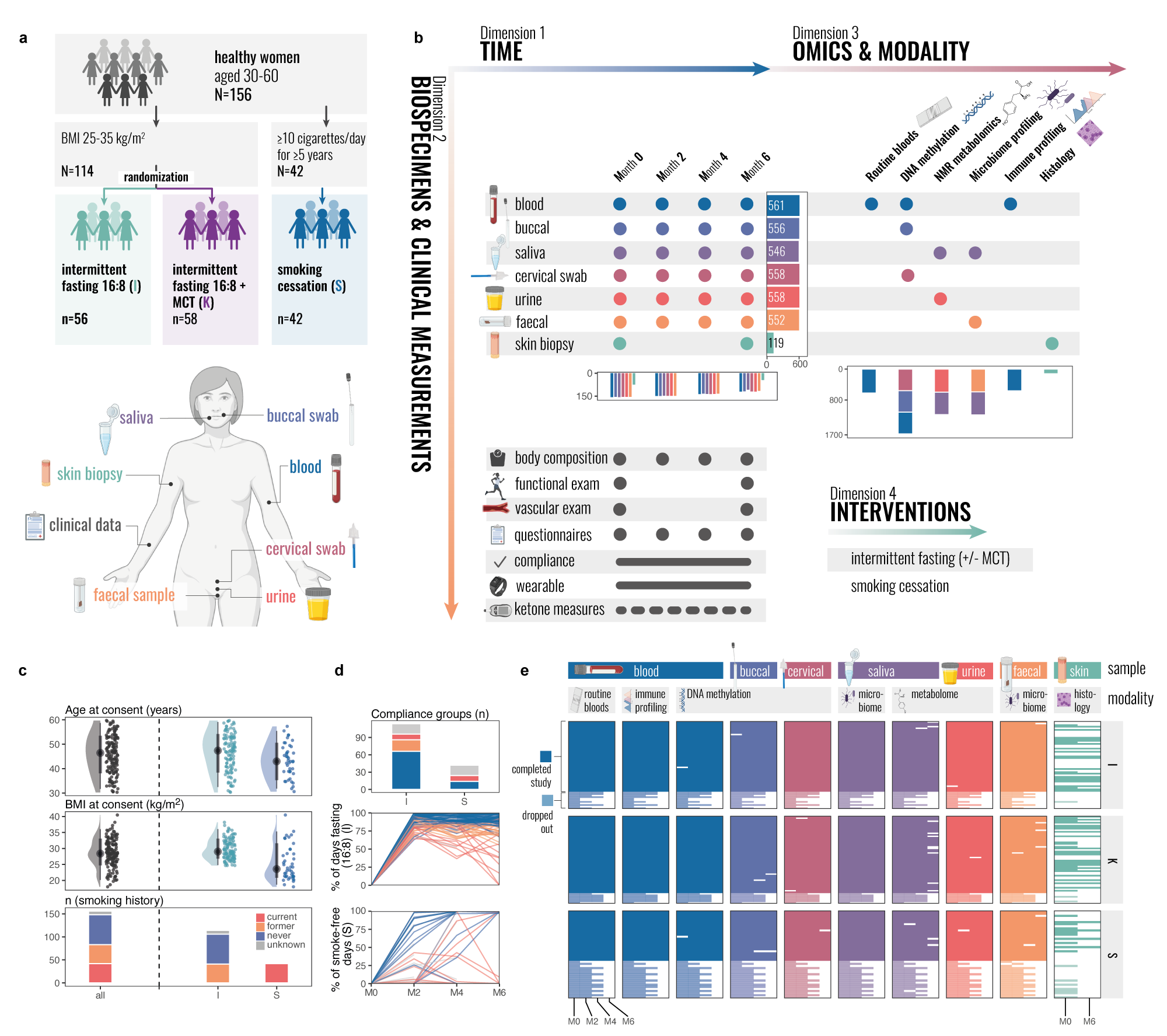 Multi-modal atlas of lifestyle interventions reveals malleability of ageing-linked molecular featuresChiara MS Herzog, Charlotte Vavourakis, Elisa Redl, and 26 more authorsbioRxiv, Dec 2025
Multi-modal atlas of lifestyle interventions reveals malleability of ageing-linked molecular featuresChiara MS Herzog, Charlotte Vavourakis, Elisa Redl, and 26 more authorsbioRxiv, Dec 2025Extending human healthspan requires understanding how lifestyle interventions impact molecular systems across tissues and time. Here, we present the TirolGESUND Lifestyle Atlas (ClinicalTrials.gov: NCT05678426), a longitudinal, multi-modal resource profiling 156 healthy women (aged 30-60 years) undergoing 6-month intermittent fasting (n=114) or smoking cessation (n=42) interventions. Participants were sampled up to four times across seven tissues and fluids, generating >3,450 biospecimens with harmonised DNA methylation, metabolomics, microbiome, and immune profiling, alongside skin histology, barrier measurements, and rich clinical metadata. We demonstrate the utility of this dataset through: (i) multi-omics-wide association studies linking traits to molecular features; (ii) integrative factor modelling revealing coordinated cross-tissue signatures; (iii) epigenetic-biomarker cross-omic associations, and (iv) CpG-level variance decomposition mapping stable, individual-specific, tissue-restricted, and intervention-responsive methylation patterns. We further show that ageing-linked features are selectively malleable: highly compliant intermittent fasting participants exhibited attenuated or even age-opposing molecular trajectories within six months. The atlas enables unprecedented within-cohort comparisons across omic layers and tissues, supporting discovery of context-dependent biomarkers, cross-system coordination, and intervention responsiveness. Data are available via an interactive portal, with sensitive data under controlled access (https://eutops.github.io/lifestyle-atlas/). This resource provides a foundation for exploring biomarker association and multi-tissue epigenetics, enabling hypothesis generation and benchmarking for systems biology and human healthspan research. Multi-tissue multi-modal profiling of two clinically-relevant lifestyle interventions in 156 women aged 30-60 Intermittent fasting modifies ageing-linked molecular features in an age-opposing direction Cross-tissue epigenetic biomarker mapping links immunity, metabolism, and microbiome Tissue-specific epigenetic plasticity maps reveal candidate meQTLs, Data sharing and interactive portal enable broad reuse for hypothesis generation and exploration
-
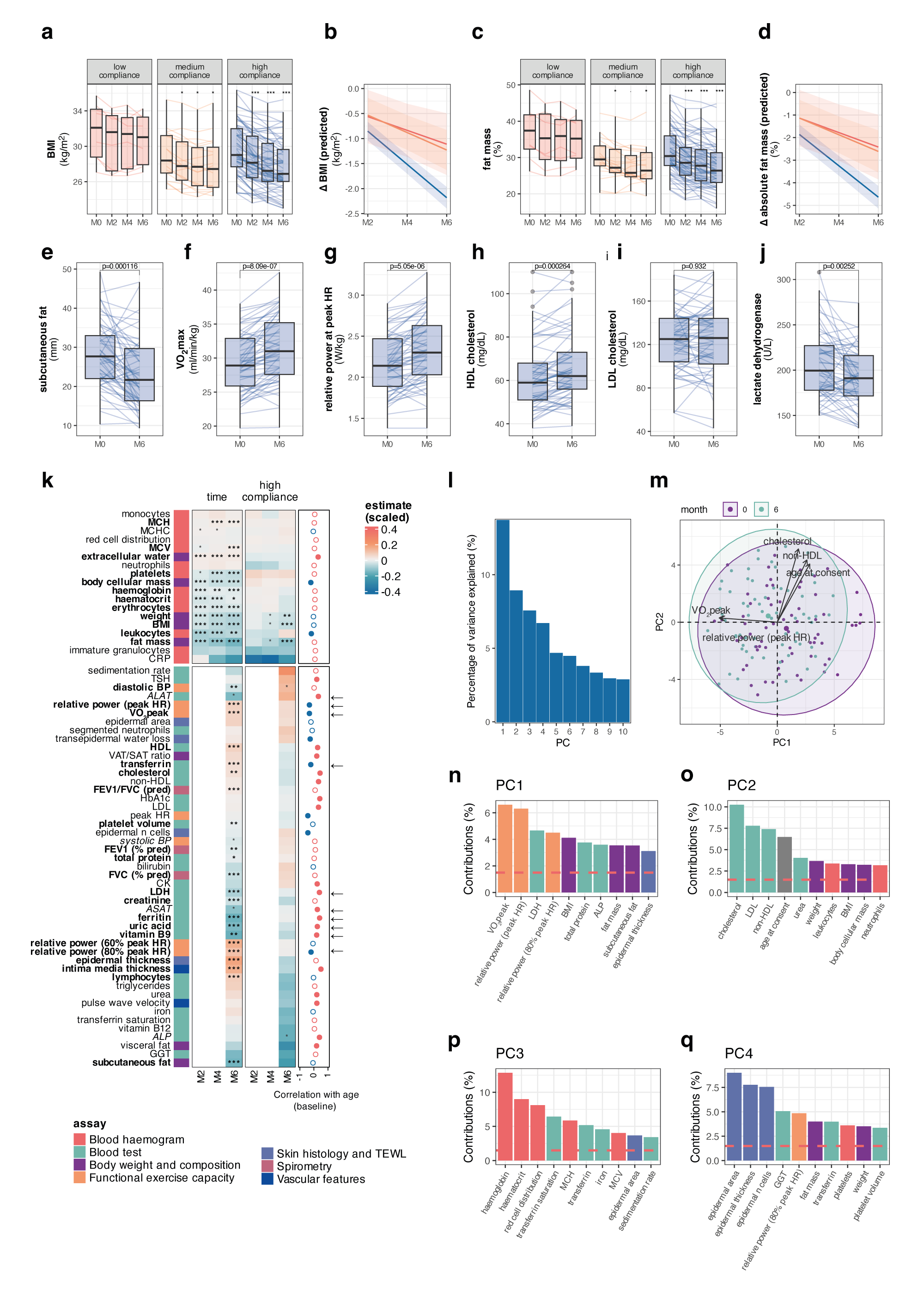 Systemic multi-omic remodelling underlies health benefits of intermittent fastingChiara Maria Stella Herzog, Charlotte D. Vavourakis, Bente Theeuwes, and 24 more authorsbioRxiv, Dec 2025
Systemic multi-omic remodelling underlies health benefits of intermittent fastingChiara Maria Stella Herzog, Charlotte D. Vavourakis, Bente Theeuwes, and 24 more authorsbioRxiv, Dec 2025While intermittent fasting (IF) promotes longevity in animal models, its systemic effects in humans remain poorly understood. Here, we present a six-month longitudinal IF intervention in 114 women (BMI 25-35) with deep clinical, molecular, and microbiome profiling across >3,400 biospecimens from six tissues. Analyses spanning >2,200 multi-omic features and 11,000 microbial function predictions demonstrate coordinated clinical benefits, including improvements in body composition and cardiorespiratory fitness, and reveal coordinated molecular responses across tissues. Iron metabolism emerged as a central axis: transferrin increased while ferritin, haemoglobin, and erythrocytes decreased, changes that opposed ageing trajectories yet remained within physiological limits. Epithelial DNA methylation biomarkers (cervical, buccal) of cancer risk reduced, while blood clocks were largely unresponsive, underscoring tissue-specificity of the epigenome. Immune profiling uncovered dynamic, partially reversible shifts. Notably, we derived a new immunophenotyping-based ImmuneAge score that increased during fasting and tracked with inflammatory function, while the pro-inflammatory cytokine IL-17A declined selectively in postmenopausal women. Oral microbiota showed rapid restructuring, whereas gut microbiota shifted more subtly toward enhanced metabolic capacity. Together, these data provide unprecedented insight into the systemic and tissue-specific responses to IF in humans and identify iron homeostasis and immune remodelling as candidate mechanisms. Our findings are available through the Lifestyle Atlas (https://eutops.github.io/lifestyle-atlas). Intermittent fasting remodels iron metabolism, opposing age-associated trajectories Epithelial but not blood methylation biomarkers respond to fasting Fasting transiently elevates ImmuneAge and TNF-producing cytotoxic T cells Oral microbiota restructure rapidly, while gut microbiota show subtler functional shifts Integrative networks link iron, immunity, adiposity, and epithelial barrier function
-
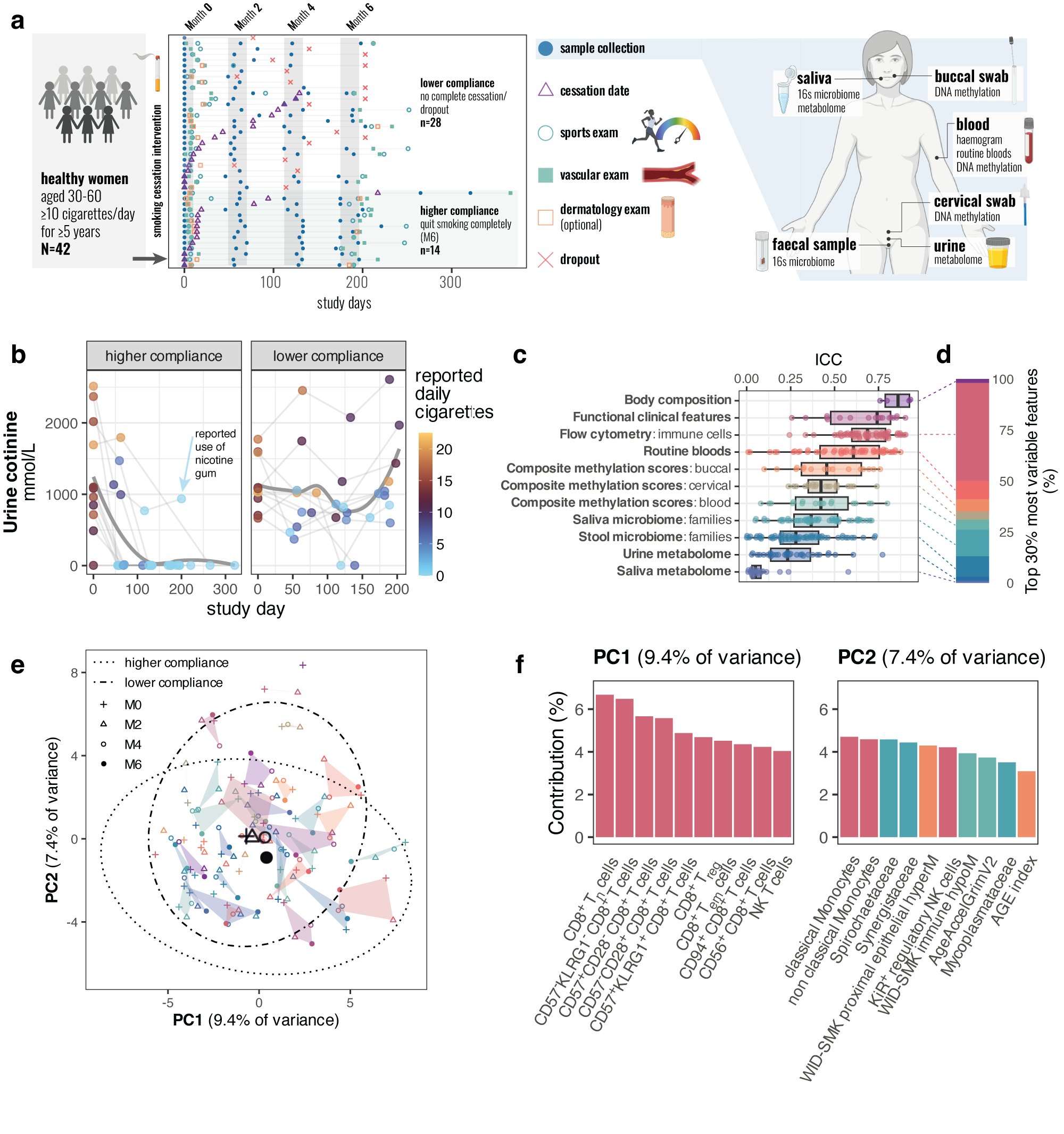 Longitudinal multi-omic evaluation of biomarkers of health and ageing over smoking cessation interventionChiara Maria Stella Herzog, Charlotte D. Vavourakis, Bente Theeuwes, and 23 more authorsbioRxiv, Dec 2025
Longitudinal multi-omic evaluation of biomarkers of health and ageing over smoking cessation interventionChiara Maria Stella Herzog, Charlotte D. Vavourakis, Bente Theeuwes, and 23 more authorsbioRxiv, Dec 2025Smoking is one of the single most important preventable risk factors for cancer and other adverse health outcomes 1,2. Smoking cessation represents a key public health intervention with the potential to reduce its negative health outcomes 2–4. While epidemiological, cross-sectional, and individual longitudinal ‘omic’ or biomarker studies have evaluated the impact of smoking cessation, no study to date has systematically profiled molecular and clinical changes in several organ systems or tissues longitudinally over the course of smoking cessation that could allow for more detailed assessment of response biomarkers and the identification of interindividual differences in the recovery of physiological functions. Here, we report the first human longitudinal multi-omic study of smoking cessation, evaluating 2,501 unique single or composite features from 1,094 longitudinal samples. Our comprehensive analysis, leveraging over half a million longitudinal data points, revealed a profound effect of smoking cessation on epigenetic biomarkers and microbiome features across multiple organ systems within 6 months of smoking cessation, alongside shifts in the immune and blood oxygenation system. Moreover, our multi-omic analysis provided unprecedented granularity that allows for identification of new cross-ome associations for mechanistic discovery. We anticipate that data and an interactive app from the Tyrol Lifestyle Atlas (eutops.github.io/lifestyle-atlas), comprising the current study and a parallel study arm evaluating the impact of diet on biomarkers of health and disease, will provide the basis for future discovery, biomarker benchmarking in their responsiveness to health-promoting interventions, and study of individualised response group, representing a major advance for personalised health monitoring using biomarkers.
-
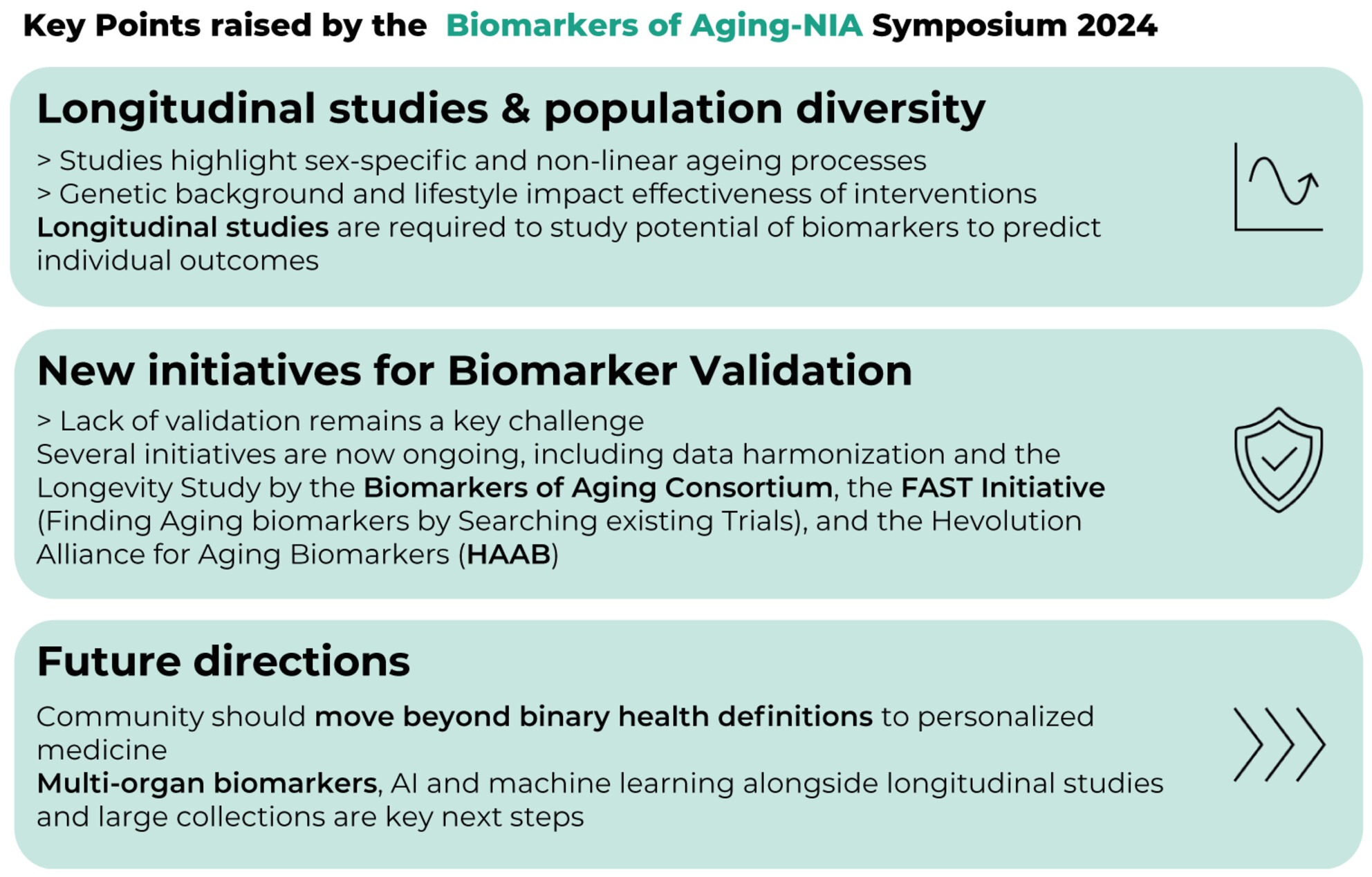 Biomarkers of Aging–NIA Joint Symposium 2024: New Insights Into Aging BiomarkersChiara Herzog, Jesse R. Poganik, Nir Barzilai, and 13 more authorsAging Cell, Dec 2025
Biomarkers of Aging–NIA Joint Symposium 2024: New Insights Into Aging BiomarkersChiara Herzog, Jesse R. Poganik, Nir Barzilai, and 13 more authorsAging Cell, Dec 2025The second Biomarkers of Aging Symposium, jointly hosted by the National Institute on Aging (NIA) Intramural Research Program and the Biomarkers of Aging Consortium (BAC) on September 12, 2024, in Baltimore, MD, convened leading researchers, clinicians, and stakeholders in the aging field to share new developments and discuss roadmaps to advance biomarkers of aging. This meeting report summarizes the highlights of this symposium and underscores the urgent need to understand longitudinal, complex, and heterogeneous processes of aging to unlock the full potential of aging biomarkers. Summary of key points raised by the Biomarkers of Aging–NIA Symposium 2024.
-
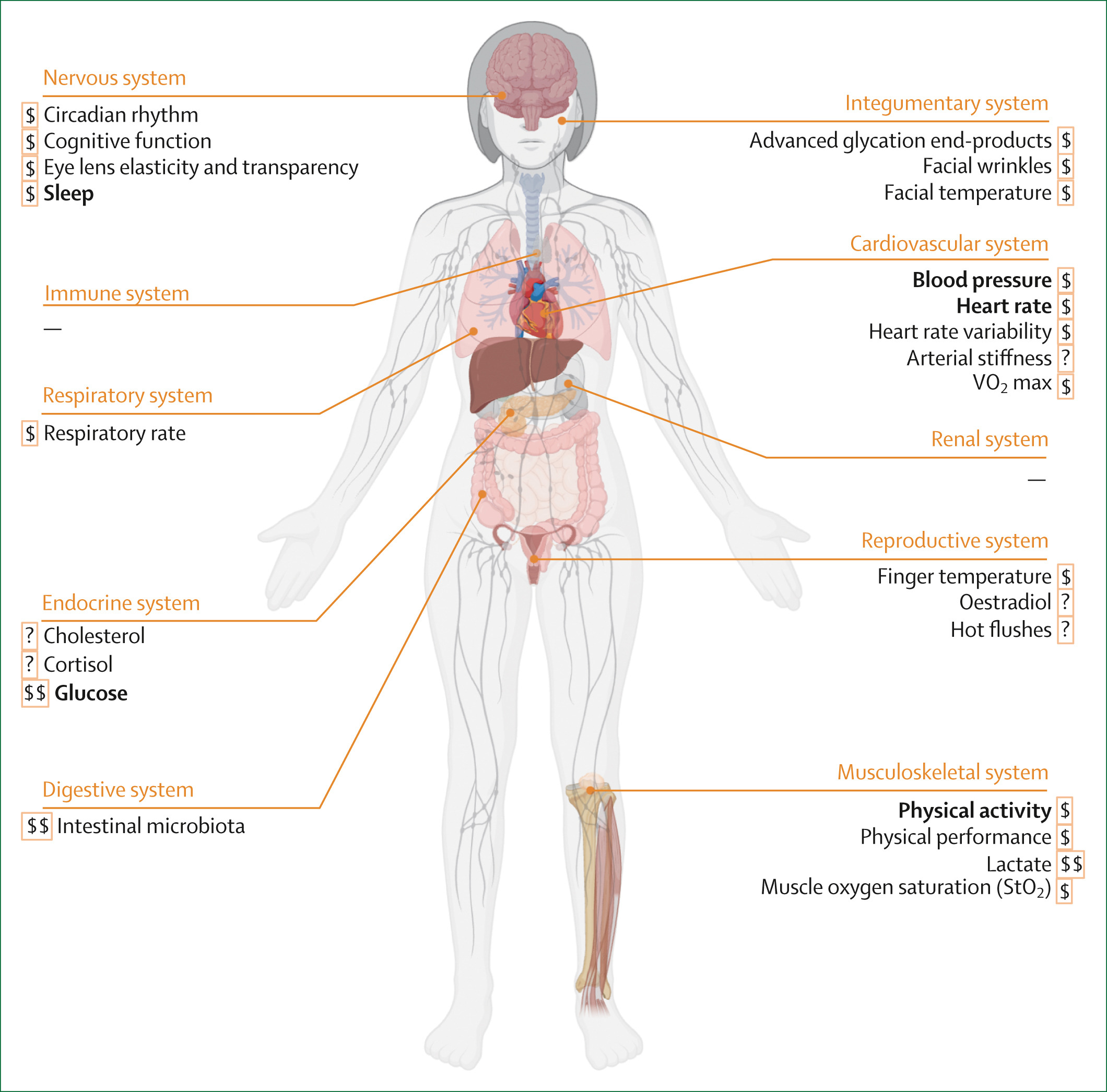 Digital biomarkers of ageing for monitoring physiological systems in community-dwelling adultsJessica K Lu, Weilan Wang, Muhammad Daniel Azlan Mahadzir, and 18 more authorsThe Lancet Healthy Longevity, Dec 2025
Digital biomarkers of ageing for monitoring physiological systems in community-dwelling adultsJessica K Lu, Weilan Wang, Muhammad Daniel Azlan Mahadzir, and 18 more authorsThe Lancet Healthy Longevity, Dec 2025Digital health technologies are transforming health care and personal health management by providing quantifiable data on physiological, behavioural, and environmental health parameters using digital biomarkers. This narrative review classified, characterised, and evaluated digital biomarkers of ageing across ten physiological systems to explore the applications of these biomarkers in research and clinical practice. The systematic search identified minimally invasive or non-invasively measured digital biomarkers suitable for longitudinal studies and practical use by community-dwelling adults. The digital biomarkers were classified according to their physiological system, characterised by their capture methods, and evaluated based on the following criteria: validity (age-associated, function-associated, and mortality-associated), generalisability, responsiveness to interventions, associations with clinical outcomes, and cost-effectiveness in large-scale settings. Digital biomarkers of ageing were found across eight physiological systems. Registered clinical trials that used these digital biomarkers as outcomes were also identified. Continued research and technological advancements are crucial for maximising the potential of digital biomarkers in promoting healthy ageing and longevity.
-
 Invigorating discovery and clinical translation of aging biomarkersErik Jacques, Chiara Herzog, Kejun Ying, and 43 more authorsNature Aging, Dec 2025
Invigorating discovery and clinical translation of aging biomarkersErik Jacques, Chiara Herzog, Kejun Ying, and 43 more authorsNature Aging, Dec 2025On 1–2 November 2024, the annual Biomarkers of Aging conference welcomed academic and industry scientists, and partners from governmental and nongovernmental organizations, to Harvard Medical School in Boston, USA, to discuss new insights into measuring and monitoring human aging, with the aim of clinical translation. In this Meeting Report, we summarize the conference and offer potential future directions for the Biomarkers of Aging Consortium and the longevity science community at large.
-
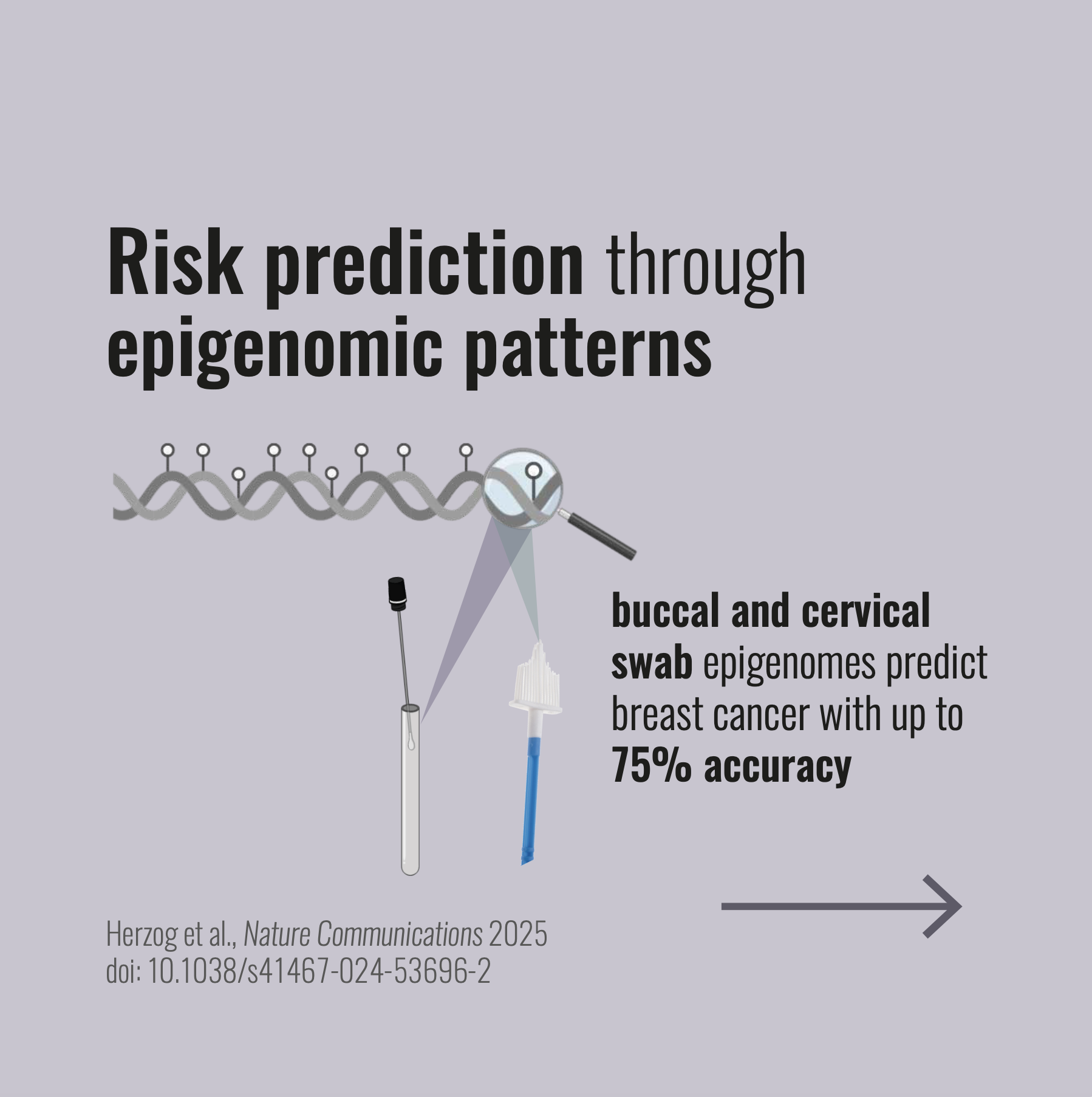 Systems epigenetic approach towards non-invasive breast cancer detectionChiara Herzog, Bente Theeuwes, Allison Jones, and 8 more authorsNature Communications, Dec 2025
Systems epigenetic approach towards non-invasive breast cancer detectionChiara Herzog, Bente Theeuwes, Allison Jones, and 8 more authorsNature Communications, Dec 2025No study has systematically compared the suitability of DNA methylation (DNAme) profiles in non-invasive samples for the detection of breast cancer (BC). We assess non-tumour DNAme in 1,100 cervical, buccal, and blood samples from BC cases and controls and find that cervical samples exhibit the largest nuber of differentially methylated sites, followed by buccal samples. No sites were significant in blood after FDR adjustment. Deriving DNAme-based classifiers for BC detection in each sample type (WID-buccal-, cervical-, or blood-BC), we achieve validation AUCs of 0.75, 0.66, and 0.51, respectively. Buccal and cervical BC-associated DNAme alterations distinguish between BC cases and controls in both surrogate and breast tissue (AUC > 0.88), yet individual sites and the directionality of methylation changes are not identical between these two sample types, and buccal sample DNAme aligns with breast methylation changes more closely. Pending additional validation, these insights may have the potential to improve non-invasive personalized BC prevention.
-
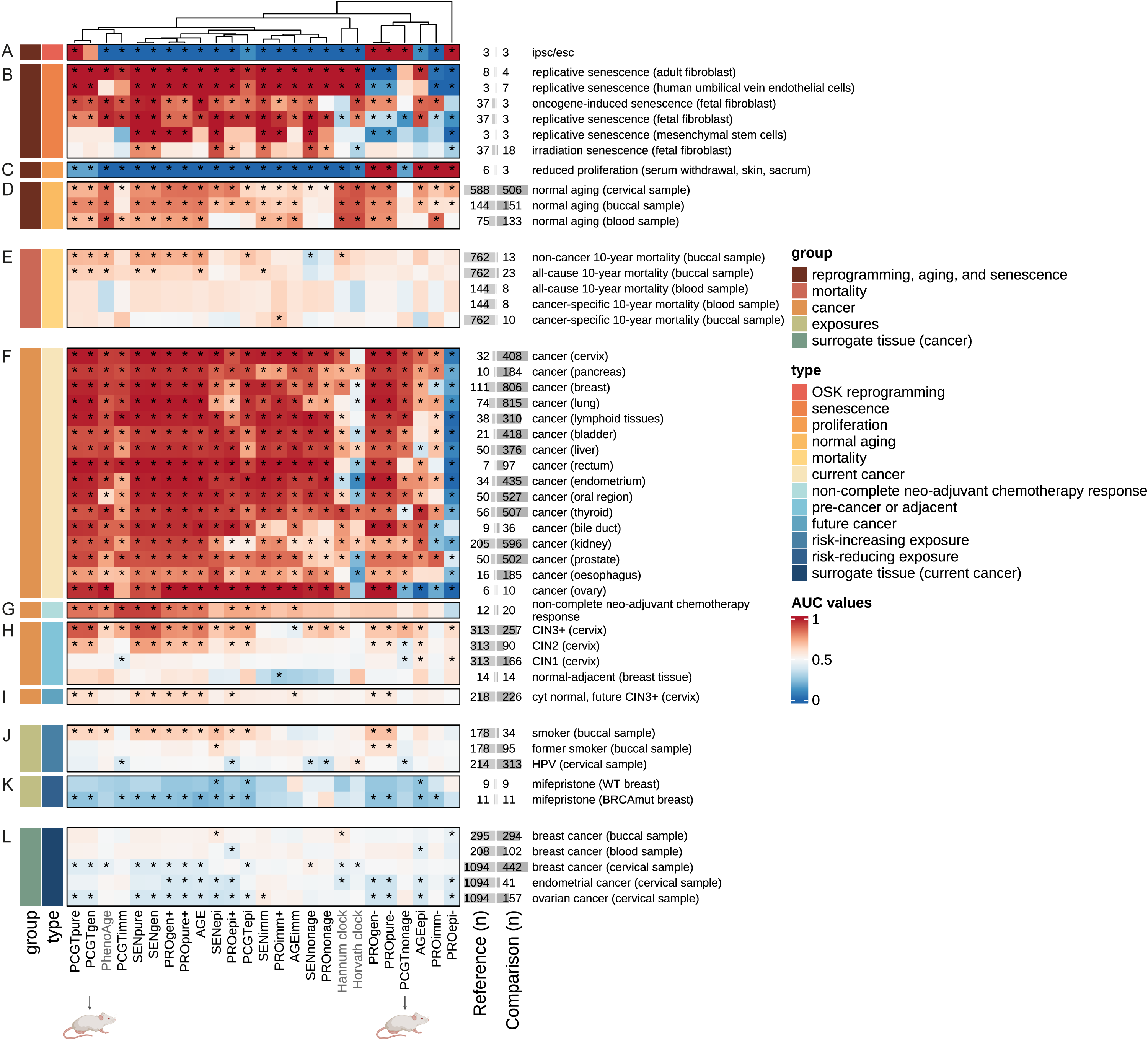 Functionally enriched epigenetic clocks reveal tissue-specific discordant aging patterns in individuals with cancerChiara Herzog, Elisa Redl, James Barrett, and 6 more authorsCommunications Medicine, Dec 2025
Functionally enriched epigenetic clocks reveal tissue-specific discordant aging patterns in individuals with cancerChiara Herzog, Elisa Redl, James Barrett, and 6 more authorsCommunications Medicine, Dec 2025Aging is a key risk factor for many diseases, including cancer, and a better understanding of its underlying molecular mechanisms may help to prevent, delay, or treat age-related pathologies. Epigenetic alterations such as DNA methylation (DNAme) changes are a hallmark of aging and form the basis of so-called epigenetic clocks, yet their functional relevance and directionality in different organs during disease development is often unclear. Here, we link cell-specific age-related DNAme changes with three key hallmarks of aging and cancer (senescence, promoter methylation in genes associated with stem cell fate, and dysregulated proliferation) to comprehensively dissect their association with current and future cancer development, carcinogen exposure or preventive measures, and mortality using data in different organs from over 12,510 human and 105 mouse samples, benchmarking against existing epigenetic clocks. Our findings offer insights into the association of functionally enriched groups of age-related DNAme changes with cancer, identify sites perturbed earliest during carcinogenesis, as well as those distinct between cancer and reprogramming that could inform strategies to prevent teratoma formation upon in vivo reprogramming. Surprisingly, both mouse and human data reveal accelerated aging in breast cancer tissue but decelerated epigenetic aging in some non-cancer surrogate samples from breast cancer patients, in particular cervical samples. This work provides evidence for discordant systemic tissue aging in breast cancer.
-
 Epigenetic signatures in surrogate tissues are able to assess cancer risk and indicate the efficacy of preventive measuresJames E. Barrett*, Chiara Herzog*, Sepideh Aminzadeh-Gohari*, and 14 more authorsCommunications Medicine, Dec 2025
Epigenetic signatures in surrogate tissues are able to assess cancer risk and indicate the efficacy of preventive measuresJames E. Barrett*, Chiara Herzog*, Sepideh Aminzadeh-Gohari*, and 14 more authorsCommunications Medicine, Dec 2025In order to advance personalized primary cancer prevention, surrogate endpoint biomarkers in distant, easy to access tissues (i.e., field defect indicators) reflecting field cancerization in the organ at risk are essential. Here we utilized medroxyprogesterone acetate and 7,12-dimethylbenzanthracene to induce mammary gland cancers in mice. We assessed epigenetic signatures reflective of carcinogen exposure, cell-type composition, mitotic age, and methylation at progesterone receptor binding sites in both, the tissue at risk (normal mammary gland; field cancerization) and distant non-at-risk organs (cervix, oviduct, and blood; field defect indicators), in mice that did and did not develop mammary gland cancers. We demonstrate that the anti-progestine mifepristone reduces the cancer risk by more than 50%. Importantly, the reduction in cancer risk is accompanied by a decline in both field cancerization and field defect indicators; specifically, epigenetic signatures in the cervix are predictive of mammary cancer formation but show tissue-specific directionality. These data encourage further exploration of epigenetic biomarkers in certain field defect-indicating tissues with a view to monitor the efficacy of cancer prevention strategies in humans.
-
 Validation of blood-based detection of breast cancer highlights importance for cross-population validationBente Theeuwes, Srikant Ambatipudi, Zdenko Herceg, and 2 more authorsNature Communications, Dec 2025
Validation of blood-based detection of breast cancer highlights importance for cross-population validationBente Theeuwes, Srikant Ambatipudi, Zdenko Herceg, and 2 more authorsNature Communications, Dec 2025 -
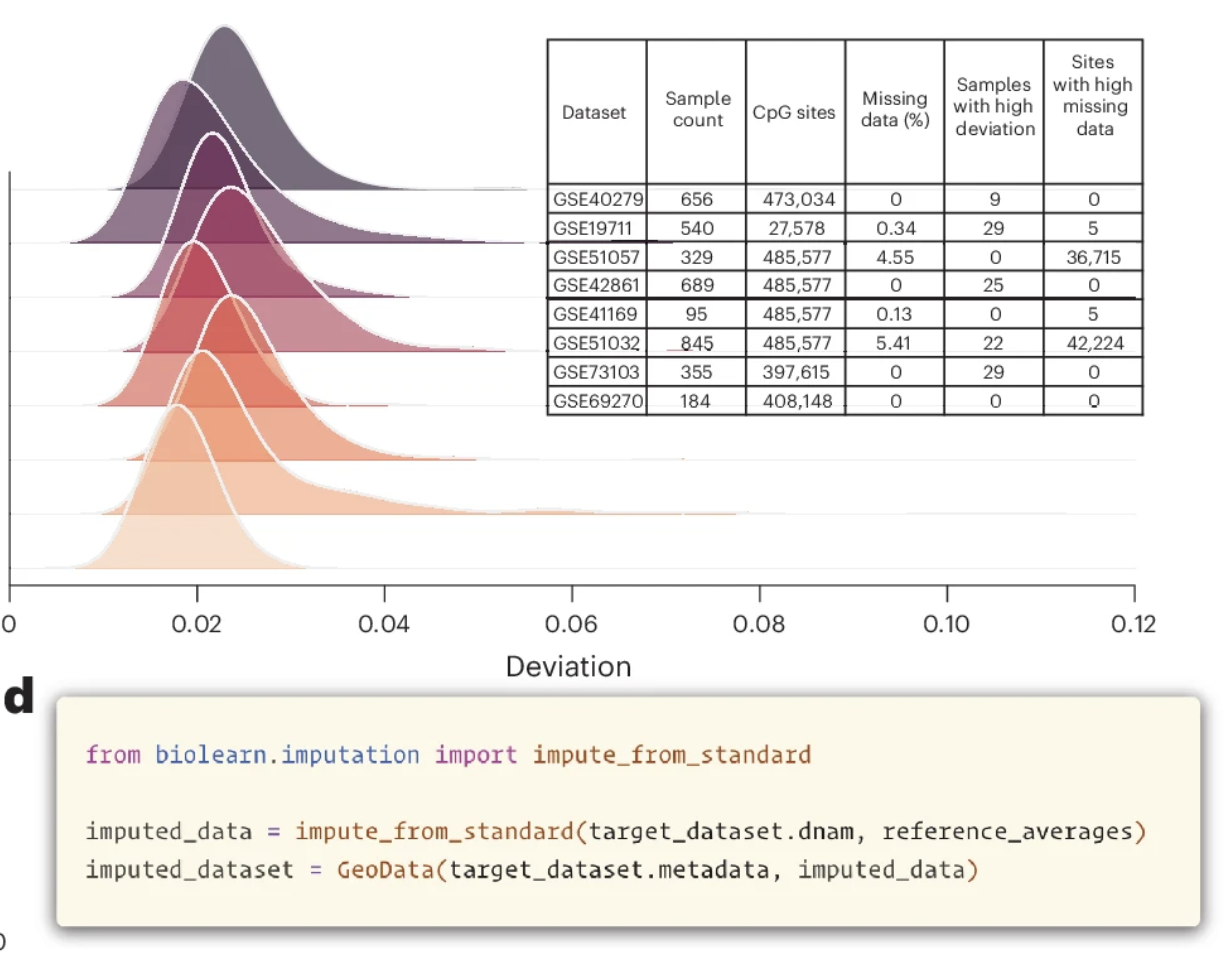 A unified framework for systematic curation and evaluation of aging biomarkersKejun Ying, Seth Paulson, Alec Eames, and 23 more authorsNature Aging, Dec 2025
A unified framework for systematic curation and evaluation of aging biomarkersKejun Ying, Seth Paulson, Alec Eames, and 23 more authorsNature Aging, Dec 2025Aging biomarkers are essential tools for quantifying biological aging, but systematic validation has been hindered by methodological inconsistencies and fragmented datasets. Here we show that the ability of traditional aging clocks to predict chronological age does not correlate with mortality prediction capacity (R = 0.12, P = 0.67), suggesting that these metrics capture distinct biological processes. We developed Biolearn, an open-source framework enabling standardized evaluation of 39 biomarkers across over 20,000 individuals from diverse cohorts. The Horvath skin and blood clock achieved the highest chronological age accuracy (R2 = 0.88), while GrimAge2 demonstrated the strongest mortality association (hazard ratio = 2.57) and healthspan prediction (hazard ratio = 2.00). Our systematic evaluation reveals considerable heterogeneity in biomarker performance across different clinical outcomes, with optimal biomarkers varying according to specific application. Biolearn provides unified data processing pipelines with quality control and cell-type deconvolution capabilities, establishing a foundation for reproducible aging research and facilitating development of robust aging biomarkers.
2024
-
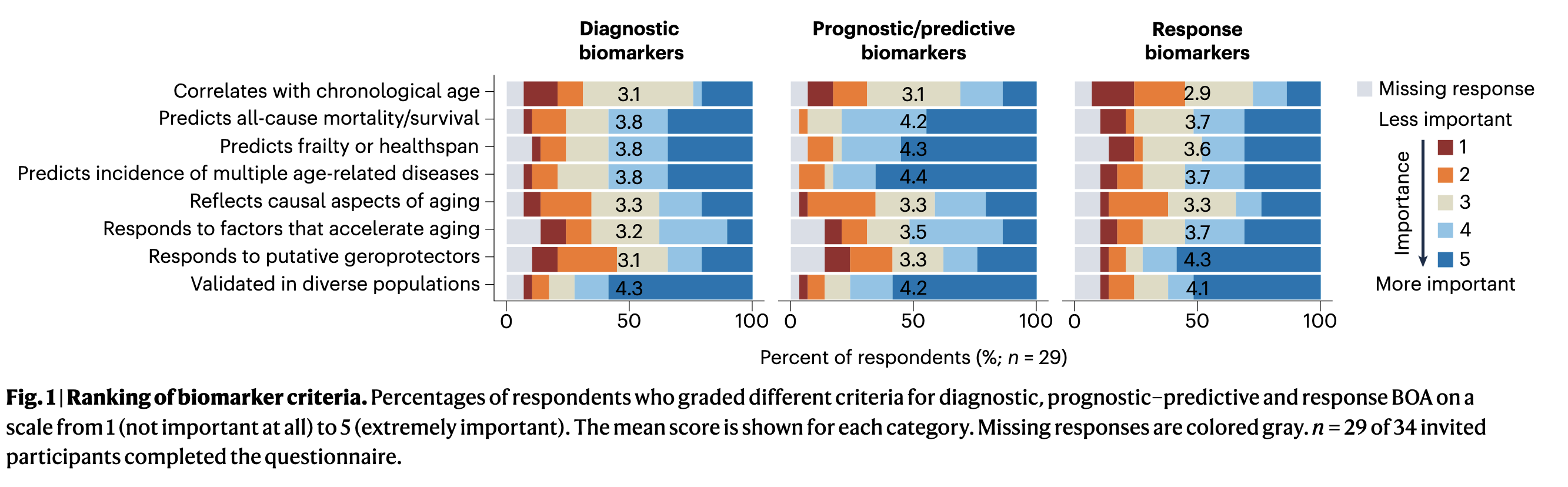 Challenges and recommendations for the translation of biomarkers of agingChiara Herzog*, Ludger J E Goeminne*, Jesse R Poganik*, and 31 more authorsNature Aging, Dec 2024
Challenges and recommendations for the translation of biomarkers of agingChiara Herzog*, Ludger J E Goeminne*, Jesse R Poganik*, and 31 more authorsNature Aging, Dec 2024Biomarkers of aging (BOA) are quantitative parameters that predict biological age and ideally its changes in response to interventions. In recent years, many promising molecular and omic BOA have emerged with an enormous potential for translational geroscience and improving healthspan. However, clinical translation remains limited, in part due to the gap between preclinical research and the application of BOA in clinical research and other translational settings. We surveyed experts in these areas to better understand current challenges for the translation of aging biomarkers. We identified six key barriers to clinical translation and developed guidance for the field to overcome them. Core recommendations include linking BOA to clinically actionable insights, improving affordability and availability to broad populations and validation of biomarkers that are robust and responsive at the level of individuals. Our work provides key insights and practical recommendations to overcome barriers impeding clinical translation of BOA. Biomarkers of aging have potential to accelerate the clinical translation of interventions that promote healthy aging but are currently limited to research. The authors identify six barriers to be overcome to enable biomarker translation, providing a roadmap to clinical implementation.
-
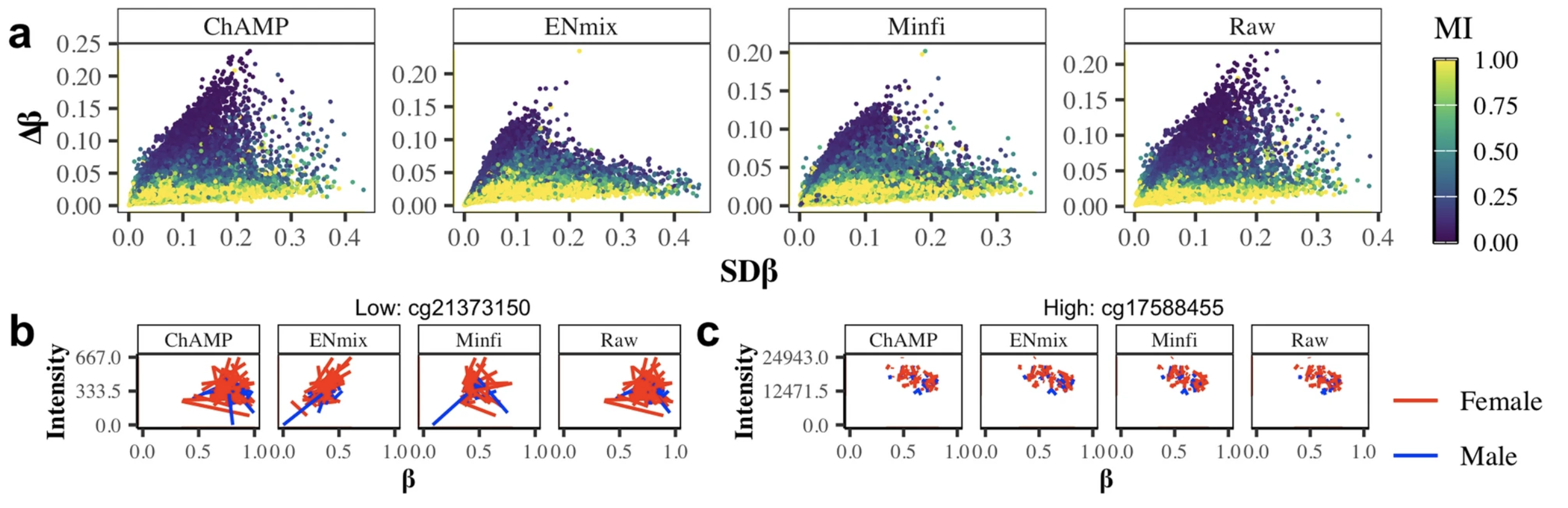 Technical and biological sources of unreliability of Infinium type II probes of the Illumina MethylationEPIC BeadChip microarrayTatiana Nazarenko, Charlotte D Vavourakis, Allison Jones, and 7 more authorsClinical Epigenetics, Dec 2024
Technical and biological sources of unreliability of Infinium type II probes of the Illumina MethylationEPIC BeadChip microarrayTatiana Nazarenko, Charlotte D Vavourakis, Allison Jones, and 7 more authorsClinical Epigenetics, Dec 2024 -
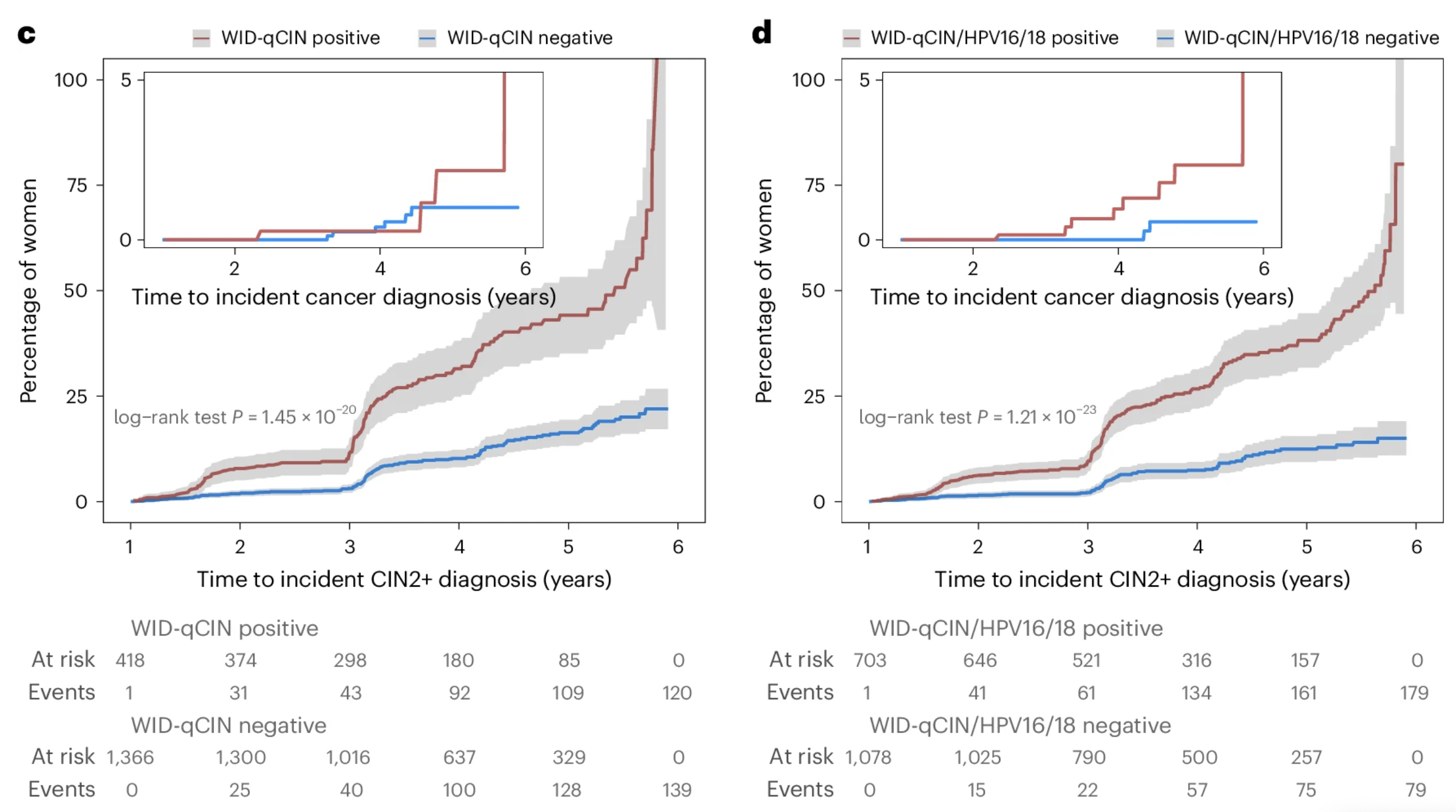 Cervical cancer screening using DNA methylation triage in a real-world populationLena Schreiberhuber, James E. Barrett, Jiangrong Wang, and 6 more authorsNature Medicine, Dec 2024
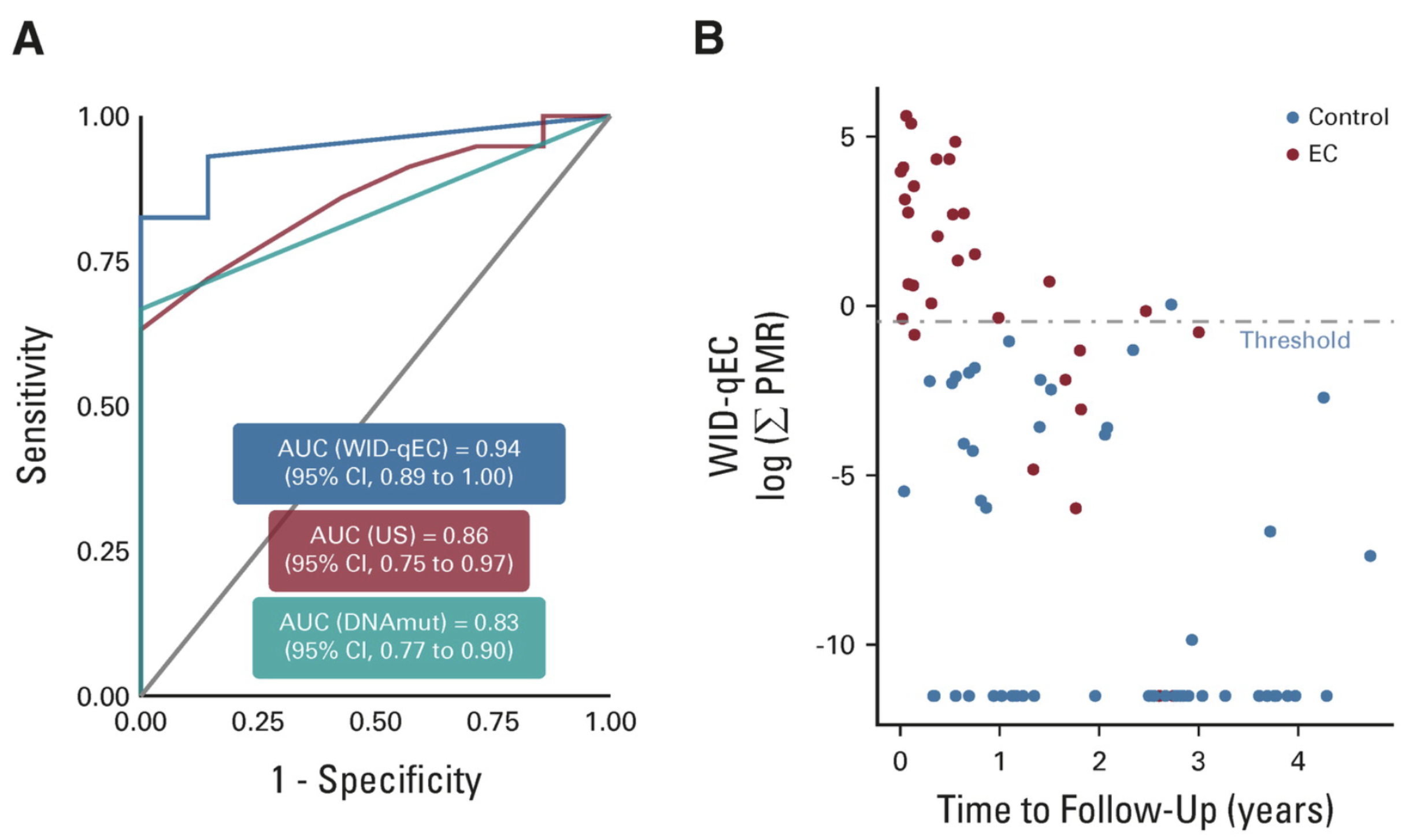
Cervical cancer screening using DNA methylation triage in a real-world populationLena Schreiberhuber, James E. Barrett, Jiangrong Wang, and 6 more authorsNature Medicine, Dec 2024Cervical cancer (CC) screening in women comprises human papillomavirus (HPV) testing followed by cytology triage of positive cases. Drawbacks, including cytology’s low reproducibility and requirement for short screening intervals, raise the need for alternative triage methods. Here we used an innovative triage technique, the WID-qCIN test, to assess the DNA methylation of human genes DPP6, RALYL and GSX1 in a real-life cohort of 28,017 women aged ≥30 years who attended CC screening in Stockholm between January and March 2017. In the analysis of all 2,377 HPV-positive samples, a combination of WID-qCIN (with a predefined threshold) and HPV16 and/or HPV18 (HPV16/18) detected 93.4% of cervical intraepithelial neoplasia grade 3 and 100% of invasive CCs. The WID-qCIN/HPV16/18 combination predicted 69.4% of incident cervical intraepithelial neoplasia grade 2 or worse compared with 18.2% predicted by cytology. Cytology or WID-qCIN/HPV16/18 triage would require 4.1 and 2.4 colposcopy referrals to detect one cervical intraepithelial neoplasia grade 2 or worse, respectively, during the 6 year period. These findings support the use of WID-qCIN/HPV16/18 as an improved triage strategy for HPV-positive women. In a real-world Swedish cohort of 28,017 women, screening for cervical cancer using a DNA methylation-based tool, the WID-qCIN test, either alone or in combination with HPV16/18 genotyping, showed better performance than cytology in triaging HPV-positive women.
-
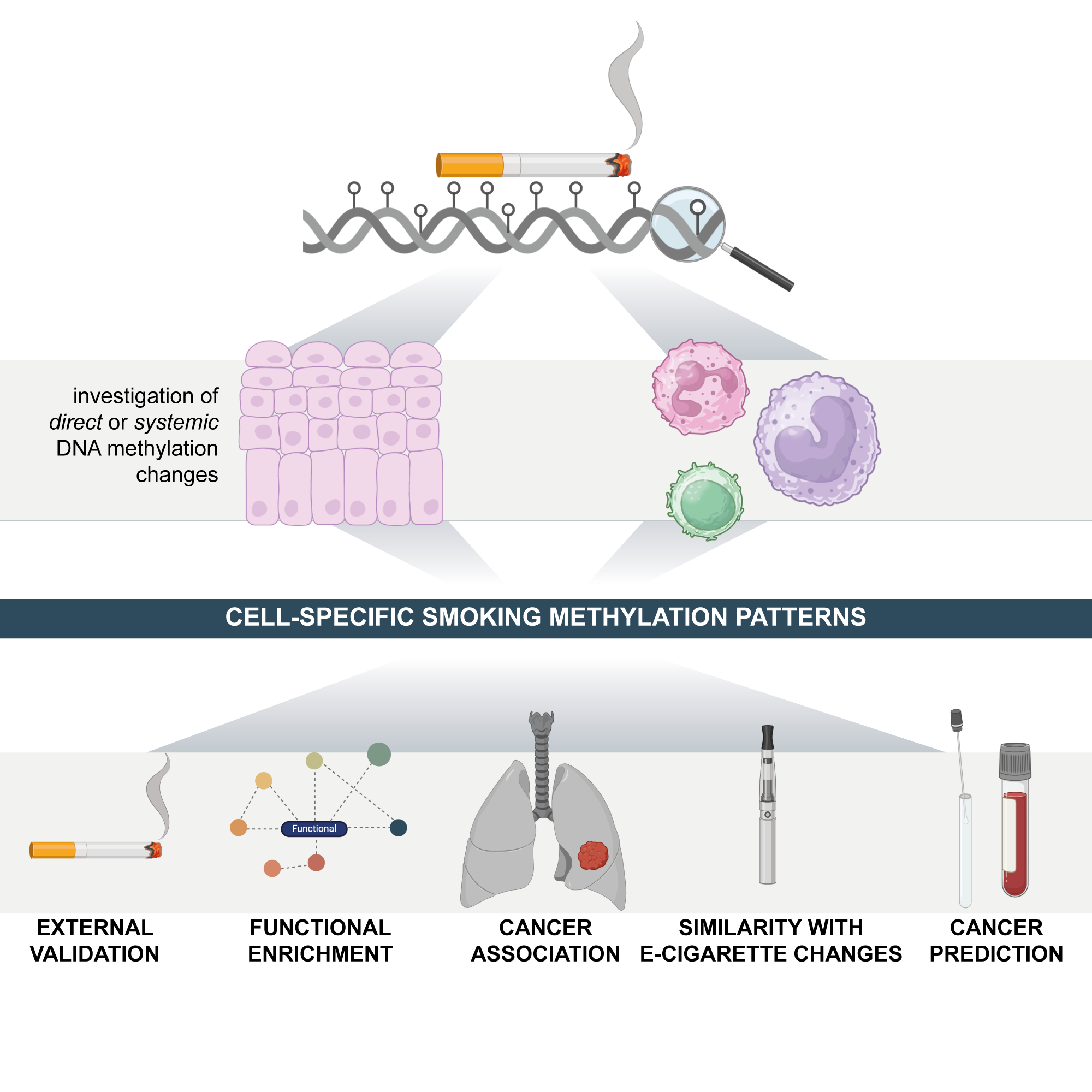 Cigarette smoking and e-cigarette use induce shared DNA methylation changes linked to carcinogenesisChiara Herzog, Allison Jones, Iona Evans, and 7 more authorsCancer Research, Dec 2024
Cigarette smoking and e-cigarette use induce shared DNA methylation changes linked to carcinogenesisChiara Herzog, Allison Jones, Iona Evans, and 7 more authorsCancer Research, Dec 2024Tobacco use is a major modifiable risk factor for adverse health outcomes, including cancer, and elicits profound epigenetic changes thought to be associated with long-term cancer risk. While electronic cigarettes (e-cigarettes) have been advocated as harm reduction alternatives to tobacco products, recent studies have revealed potential detrimental effects, highlighting the urgent need for further research into the molecular and health impacts of e-cigarettes. Here, we applied computational deconvolution methods to dissect the cell- and tissue-specific epigenetic effects of tobacco or e-cigarette use on DNA methylation (DNAme) in over 3,500 buccal/saliva, cervical, or blood samples, spanning epithelial and immune cells at directly and indirectly exposed sites. The 535 identified smoking-related DNAme loci (CpGs) clustered into four functional groups, including detoxification or growth signaling, based on cell type and anatomical site. Loci hypermethylated in buccal epithelial cells of smokers associated with NOTCH1/RUNX3/growth factor receptor signaling also exhibited elevated methylation in cancer tissue and progressing lung carcinoma in situ lesions, and hypermethylation of these sites predicted lung cancer development in buccal samples collected from smokers up to 22 years prior to diagnosis, suggesting a potential role in driving carcinogenesis. Alarmingly, these CpGs were also hypermethylated in e-cigarette users with a limited smoking history. This study sheds light on the cell type-specific changes to the epigenetic landscape induced by smoking-related products.
-
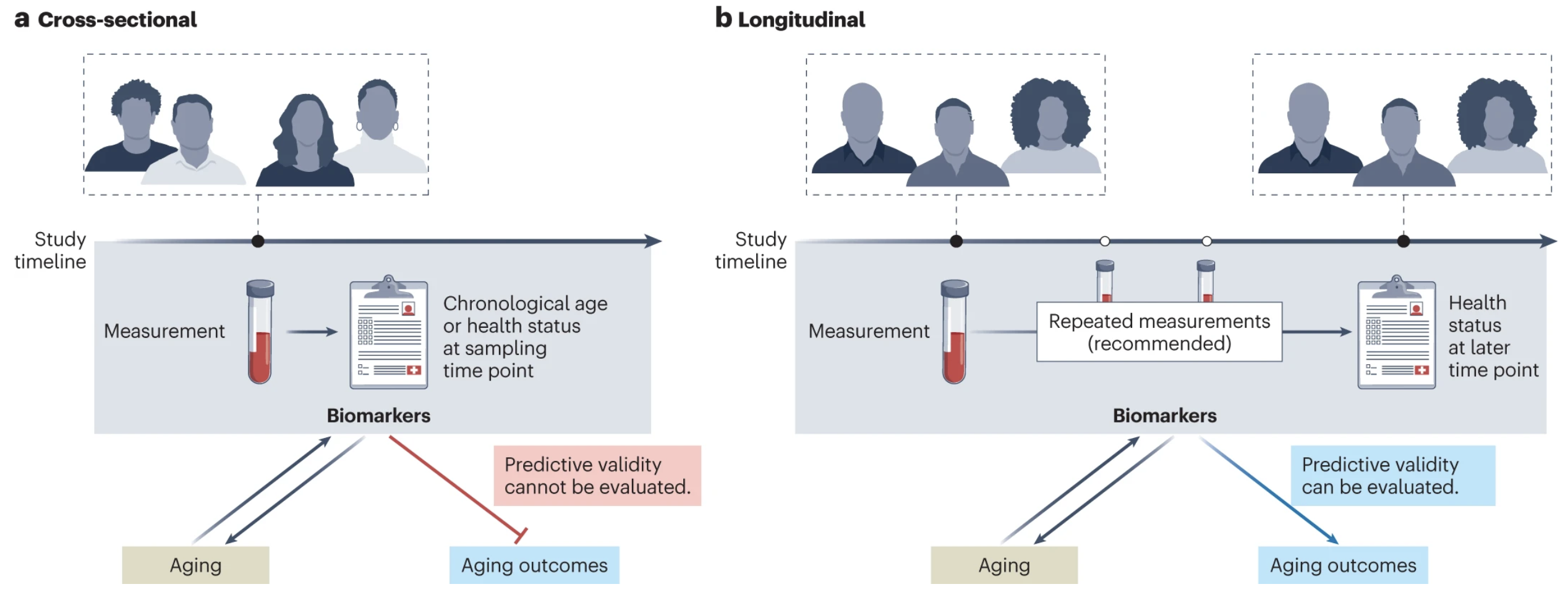 Validation of biomarkers of agingMahdi Moqri*, Chiara Herzog*, Jesse R. Poganik*, and 26 more authorsNature Medicine, Dec 2024
Validation of biomarkers of agingMahdi Moqri*, Chiara Herzog*, Jesse R. Poganik*, and 26 more authorsNature Medicine, Dec 2024The search for biomarkers that quantify biological aging (particularly ‘omic’-based biomarkers) has intensified in recent years. Such biomarkers could predict aging-related outcomes and could serve as surrogate endpoints for the evaluation of interventions promoting healthy aging and longevity. However, no consensus exists on how biomarkers of aging should be validated before their translation to the clinic. Here, we review current efforts to evaluate the predictive validity of omic biomarkers of aging in population studies, discuss challenges in comparability and generalizability and provide recommendations to facilitate future validation of biomarkers of aging. Finally, we discuss how systematic validation can accelerate clinical translation of biomarkers of aging and their use in gerotherapeutic clinical trials. Robust validation of biomarkers of aging will be critical to their clinical translation; here, authors review the key challenges and propose recommendations to overcome them.
-
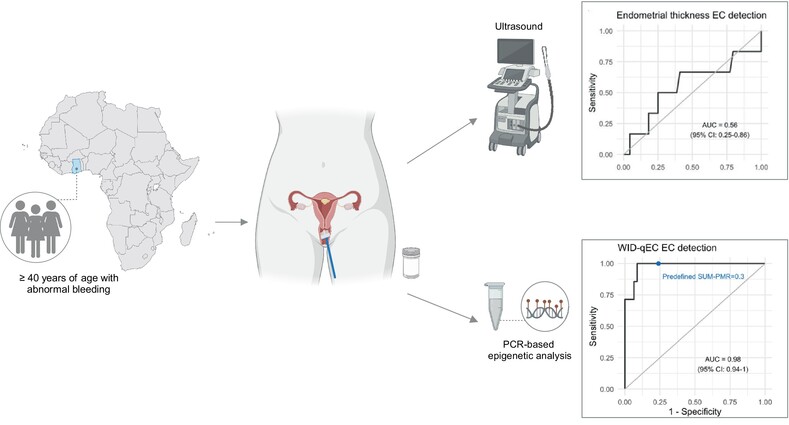 Performance of the WID‐qEC test to detect uterine cancers in black women with abnormal uterine bleeding: A prospective observational cohort study in GhanaSebastian Ken‐Amoah, Elisa Redl, Bright K. S. Domson, and 14 more authorsInternational Journal of Cancer, Dec 2024
Performance of the WID‐qEC test to detect uterine cancers in black women with abnormal uterine bleeding: A prospective observational cohort study in GhanaSebastian Ken‐Amoah, Elisa Redl, Bright K. S. Domson, and 14 more authorsInternational Journal of Cancer, Dec 2024The burden of uterine cancer is growing and, in the US and UK, mortality rates are poorest among black women. Early detection of these cancers is critical and poor performance of ultrasound in black women may contribute to adverse outcomes. Limited data on this topic are available from Africa. We assessed whether a simple DNA methylation test, the WID‐qEC, enables detection of all epithelial uterine (endometrial and cervical) cancers in women presenting with abnormal uterine bleeding (AUB) in Ghana. Among 118 women ≥40 years presenting with AUB, 106 consented to the study and a cervicovaginal sample was obtained for WID‐qEC testing. Subsequent to ultrasound assessment 102 women had a cervical or endometrial biopsy. Histopathology, ultrasound and WID‐qEC testing were analyzed and compared. Among the 102 volunteers, 8 and 15 were diagnosed with endometrial and cervical cancer (EC and CC), respectively. The receiver operating characteristic (ROC) area under the curve (AUC) was 0.56 (95% confidence interval [CI] 0.25–0.86) for sonographic endometrial thickness (ET) and 0.98 (95% CI 0.94–1.00) for the WID‐qEC test. Sensitivity and specificity of the prespecified ET ≥5 mm were 66.7% (95% CI 24.1–94.0) and 22.7 (95% CI 12.0–38.2) and for the prespecified WID‐qEC SUM‐PMR ≥ 0.3 were 100% (95% CI 56.1–100.0) and 76.1 (96%CI 60.9–86.9), respectively. In addition, 15 CCs were detected by the WID‐qEC test [sensitivity 100% (95% CI 74.7–100.0)]. The WID‐qEC test accurately detects both EC and CC in black women presenting with AUB. Endometrial cancer (EC) is increasing significantly among women in low‐ and middle‐income countries (LMICs). EC detection may be improved by WID‐qEC testing, a DNA methylation‐based approach, though its use has been limited primarily to Caucasian populations. Here, the authors investigated the ability of WID‐qEC testing to detect endometrial and cervical cancer in black women in Ghana who presented with abnormal uterine bleeding. WID‐qEC testing successfully identified 100% of endometrial and cervical cancers in the study population. Utilizing simple sample collection and low‐cost, high‐throughput PCR‐based analysis, WID‐qEC is a promising tool for EC detection for all women in LMICs.
-
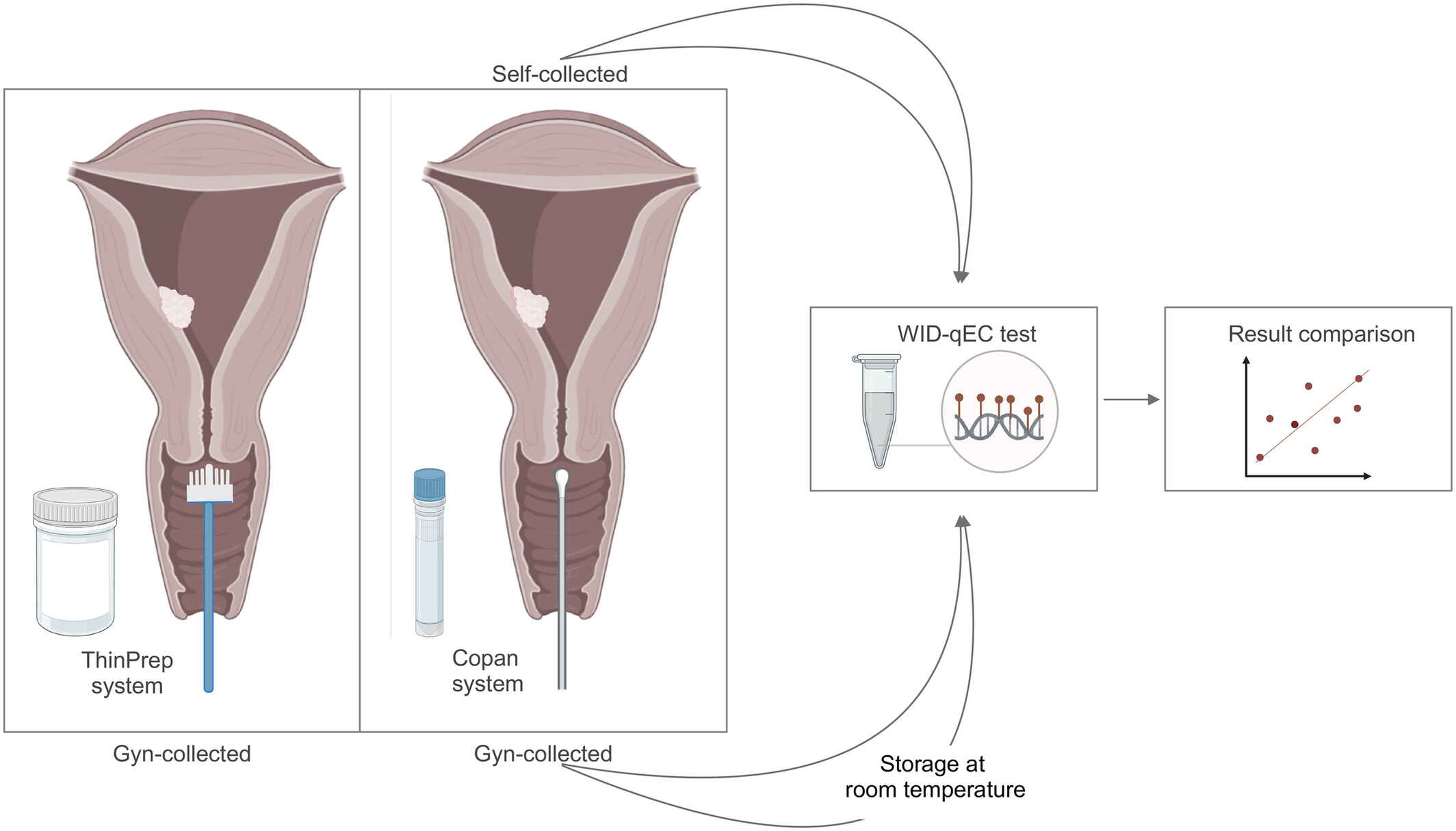 High performance of the DNA methylation‐based WID‐qEC test for detecting uterine cancers independent of sampling modalitiesOjone Illah, Malcolm Scott, Elisa Redl, and 16 more authorsInternational Journal of Cancer, Dec 2024
High performance of the DNA methylation‐based WID‐qEC test for detecting uterine cancers independent of sampling modalitiesOjone Illah, Malcolm Scott, Elisa Redl, and 16 more authorsInternational Journal of Cancer, Dec 2024Endometrial cancer (EC) is the most prevalent gynaecological cancer in high‐income countries and its incidence is continuing to rise sharply. Simple and objective tools to reliably detect women with EC are urgently needed. We recently developed and validated the DNA methylation (DNAme)‐based women’s cancer risk identification—quantitative polymerase chain reaction test for endometrial cancer (WID‐qEC) test that could address this need. Here, we demonstrate that the stability of the WID‐qEC test remains consistent regardless of: (i) the cervicovaginal collection device and sample media used (Cervex brush and PreservCyt or FLOQSwab and eNAT), (ii) the collector of the specimen (gynaecologist‐ or patient‐based), and (iii) the precise sampling site (cervical, cervicovaginal and vaginal). Furthermore, we demonstrate sample stability in eNAT medium for 7 days at room temperature, greatly facilitating the implementation of the test into diagnostic laboratory workflows. When applying FLOQSwabs (Copan) in combination with the eNAT (Copan) sample collection media, the sensitivity and specificity of the WID‐qEC test to detect uterine (i.e., endometrial and cervical) cancers in gynaecologist‐taken samples was 92.9% (95% confidence interval [CI] = 75.0%–98.8%) and 98.6% (95% CI = 91.7%–99.9%), respectively, whilst the sensitivity and specificity in patient collected self‐samples was 75.0% (95% CI = 47.4%–91.7%) and 100.0% (95% CI = 93.9%–100.0%), respectively. Taken together these data confirm the robustness and clinical potential of the WID‐qEC test. Subjective diagnostic tests with modest accuracy, such as cytology or ultrasound, are currently used to assess the likelihood of uterine cancer in women with abnormal bleeding. Here, the authors show that the DNA methylation‐based women’s cancer risk identification—quantitative polymerase chain reaction test for endometrial cancer (WID‐qEC) test they have previously developed has high sensitivity and specificity in detecting uterine cancers in symptomatic women irrespective of the sample collection device and medium, sample collector and precise sampling site. Furthermore, they demonstrate the compatibility of the WID‐qEC test used with the Copan sampling system with established diagnostic laboratory workflows, confirming the robustness and clinical potential of the WID‐qEC test.
-
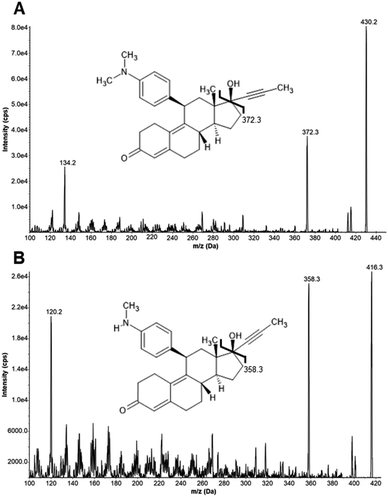 A validated HPLC-MS/MS method for the quantification of systemic mifepristone after subcutaneous application in miceJulia Tevini, Sepideh Aminzadeh-Gohari, Daniela D. Weber, and 8 more authorsAnalytical Methods, Dec 2024
A validated HPLC-MS/MS method for the quantification of systemic mifepristone after subcutaneous application in miceJulia Tevini, Sepideh Aminzadeh-Gohari, Daniela D. Weber, and 8 more authorsAnalytical Methods, Dec 2024Quantification of the antiprogestin mifepristone and its active metabolite metapristone in plasma and tissue of female mice using HPLC-MS/MS.
-
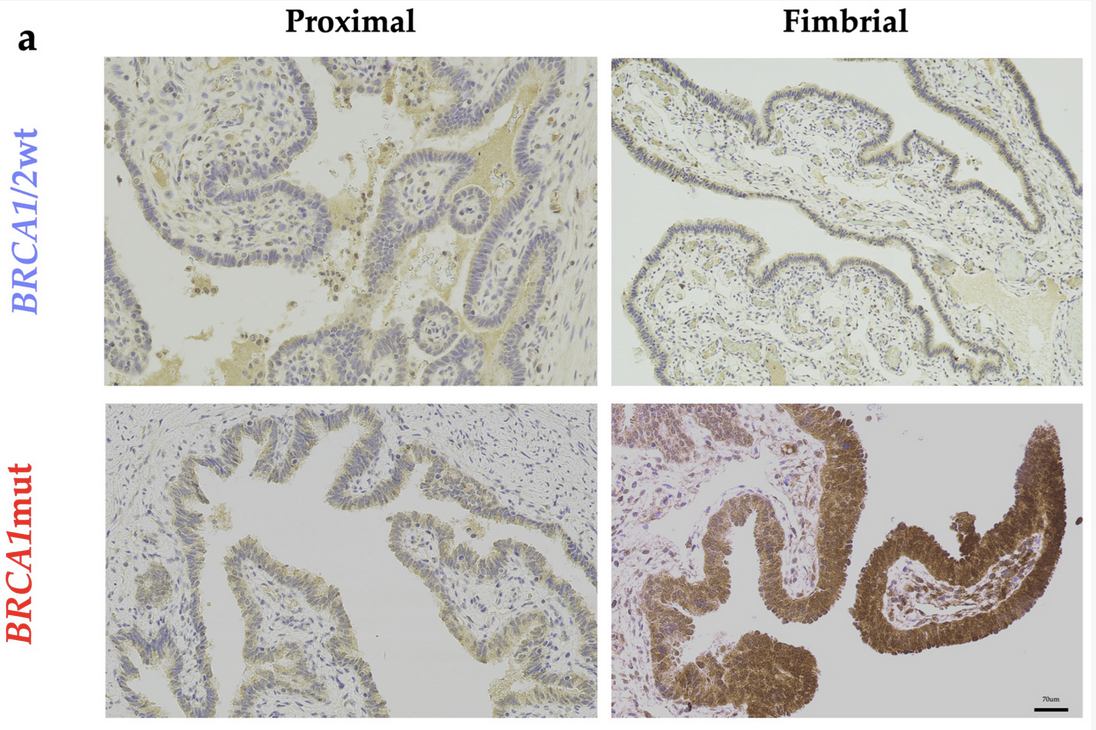 Natural Killer Cell Dysfunction in Premenopausal BRCA1 Mutation Carriers: A Potential Mechanism for Ovarian CarcinogenesisShaun Haran, Kantaraja Chindera, May Sabry, and 17 more authorsCancers, Dec 2024
Natural Killer Cell Dysfunction in Premenopausal BRCA1 Mutation Carriers: A Potential Mechanism for Ovarian CarcinogenesisShaun Haran, Kantaraja Chindera, May Sabry, and 17 more authorsCancers, Dec 2024Since the discovery of a BRCA1 mutation nearly 30 years ago, definitive methods for ovarian cancer-risk reduction has remained unchanged, requiring carriers of a pathogenic mutation to consider undergoing surgical removal of both fallopian tubes and ovaries from the ages of 35 to 40 years. The sequelae of these invasive measures include a profound impact on general health, psychological well-being, sexual and reproductive function. Identifying targetable aberrances in BRCA1 mutation carriers, such as sex hormones, or adopting mechanisms aimed at enhancing tumor immunosurveillance may facilitate the reduction of early cancer-promoting events. This is particularly relevant for the fimbrial fallopian tube, where the majority of epithelial ovarian cancers originate. Utilizing a proof-of-concept for potentially targetable aberrances may further the development of non-surgical adjuncts or surveillance methods aimed at cancer-risk reduction for women deferring or declining surgery. Background: Tissue-specificity for fimbrial fallopian tube ovarian carcinogenesis remains largely unknown in BRCA1 mutation carriers. We aimed to assess the cell autonomous and cell-nonautonomous implications of a germline BRCA1 mutation in the context of cancer immunosurveillance of CD3− CD56+ natural killer (NK) cells. Methods: Premenopausal BRCA1 mutation carriers versus age-matched non-carriers were compared. Daily urinary 5β-pregnanediol levels were used to determine progesterone metabolomics across an ovarian cycle. Using peripherally acquired NK cells the cell-mediated cytotoxicity of tumor targets (OVCAR-3, K-562) was determined using live cellular impedance (xCELLigence®) and multicolor flow cytometry. Hypoxia-inducible factor 1-alpha (HIF-1α) immunohistochemistry of cancer-free fallopian tube specimens allowed a comparison of proximal versus distal portions. Utilizing these findings the role of environmental factors relevant to the fimbrial fallopian tube (progesterone, hypoxia) on NK cell functional activity were studied in an ovarian phase-specific manner. Results: BRCA1 mutation carriers demonstrate a differential progesterone metabolome with a phase-specific reduction of peripheral NK cell functional activity. Progesterone exposure further impairs NK cell-mediated cytotoxicity in a dose-dependent manner, which is reversed with the addition of mifepristone (1.25 µM). The fimbrial fallopian tube demonstrated significantly higher HIF-1α staining, particularly in BRCA1 mutation carriers, reflecting a site-specific ‘hypoxic niche’. Exposure to hypoxic conditions (1% O2) can further impair tumor cytotoxicity in high-risk carriers. Conclusions: Phase-specific differential NK cell activity in BRCA1 mutation carriers, either systemically or locally, may favor site-specific pre-invasive carcinogenesis. These cumulative effects across a reproductive lifecycle in high-risk carriers can have a detrimental effect further supporting epidemiological evidence for ovulation inhibition.
2023
-
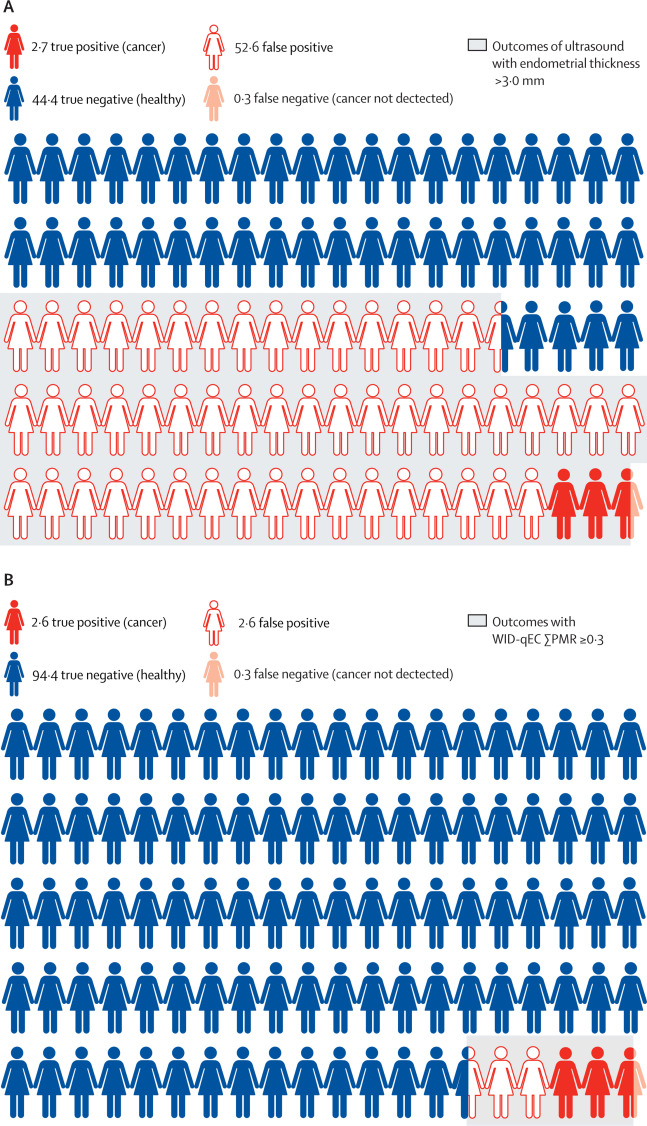 Performance of the WID-qEC test versus sonography to detect uterine cancers in women with abnormal uterine bleeding (EPI-SURE): a prospective, consecutive observational cohort study in the UKIona Evans*, Daniel Reisel*, Allison Jones*, and 11 more authorsThe Lancet Oncology, Dec 2023
Performance of the WID-qEC test versus sonography to detect uterine cancers in women with abnormal uterine bleeding (EPI-SURE): a prospective, consecutive observational cohort study in the UKIona Evans*, Daniel Reisel*, Allison Jones*, and 11 more authorsThe Lancet Oncology, Dec 2023 -
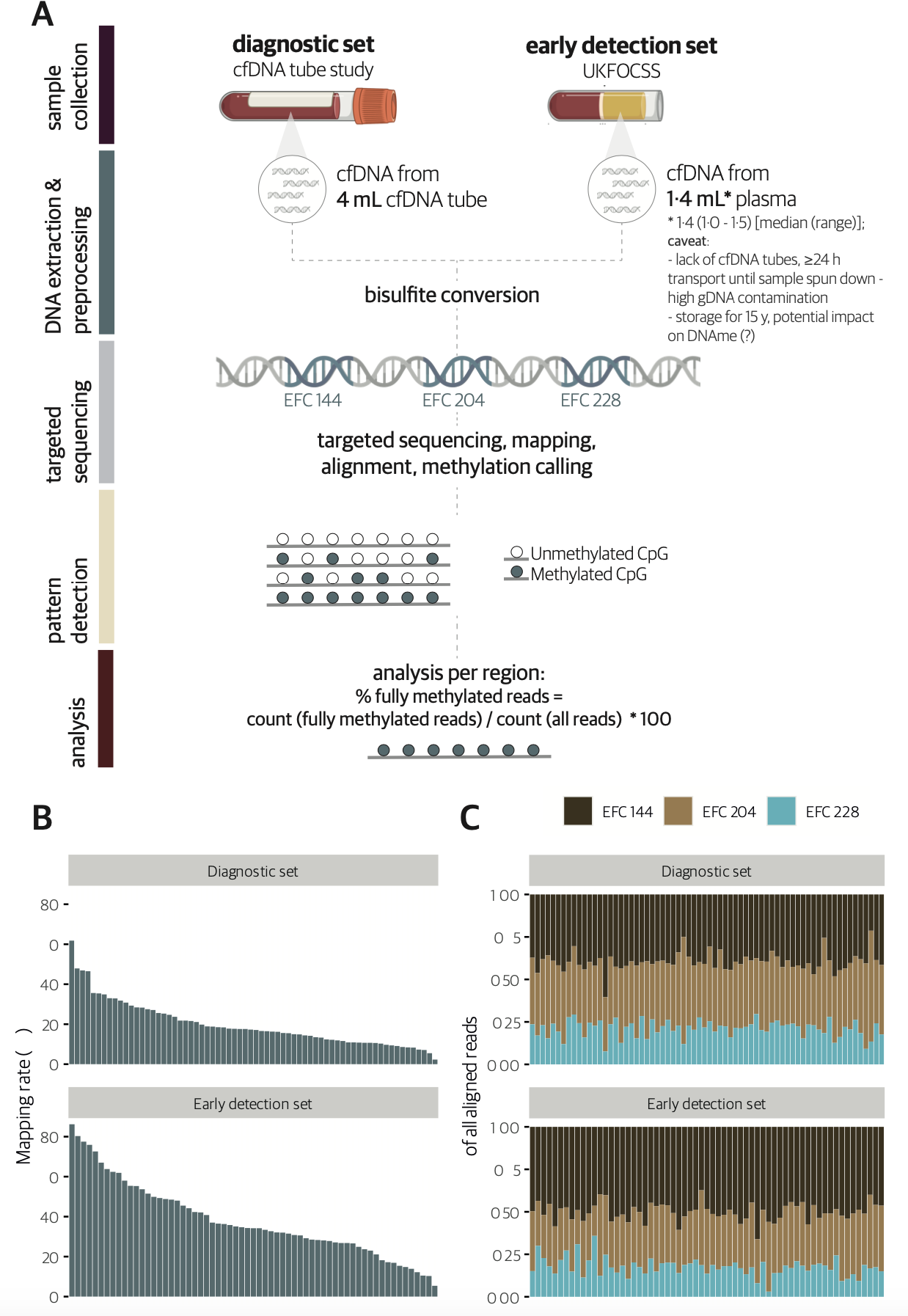 Plasma cell‐free DNA methylation analysis for ovarian cancer detection: Analysis of samples from a case‐control study and an ovarian cancer screening trialChiara Herzog, Allison Jones, Iona Evans, and 18 more authorsInternational Journal of Cancer, Dec 2023
Plasma cell‐free DNA methylation analysis for ovarian cancer detection: Analysis of samples from a case‐control study and an ovarian cancer screening trialChiara Herzog, Allison Jones, Iona Evans, and 18 more authorsInternational Journal of Cancer, Dec 2023Analysis of cell‐free DNA methylation (cfDNAme), alone or combined with CA125, could help to detect ovarian cancers earlier and may reduce mortality. We assessed cfDNAme in regions of ZNF154, C2CD4D and WNT6 via targeted bisulfite sequencing in diagnostic and early detection (preceding diagnosis) settings. Diagnostic samples were obtained via prospective blood collection in cell‐free DNA tubes in a convenience series of patients with a pelvic mass. Early detection samples were matched case‐control samples derived from the UK Familial Ovarian Cancer Screening Study (UKFOCSS). In the diagnostic set (ncases = 27, ncontrols = 41), the specificity of cfDNAme was 97.6% (95% CI: 87.1%‐99.9%). High‐risk cancers were detected with a sensitivity of 80% (56.3%‐94.3%). Combination of cfDNAme and CA125 resulted in a sensitivity of 94.4% (72.7%‐99.9%) for high‐risk cancers. Despite technical issues in the early detection set (ncases = 29, ncontrols = 29), the specificity of cfDNAme was 100% (88.1%‐100.0%). We detected 27.3% (6.0%‐61.0%) of high‐risk cases with relatively lower genomic DNA (gDNA) contamination. The sensitivity rose to 33.3% (7.5%‐70.1%) in samples taken <1 year before diagnosis. We detected ovarian cancer in several patients up to 1 year before diagnosis despite technical limitations associated with archival samples (UKFOCSS). Combined cfDNAme and CA125 assessment may improve ovarian cancer screening in high‐risk populations, but future large‐scale prospective studies will be required to validate current findings. Our findings indicate that combining cell‐free DNA methylation analysis in three genetic regions with CA125 may improve the sensitivity of ovarian cancer detection in high‐risk women. Women with a positive score in either diagnostic modality, particularly double‐positive individuals, could be referred for a PET‐CT scan to rule out a positive result due to a different cancer, and could undergo surgery even in the absence of a visible ovarian tumour on imaging given the high specificity of cfDNAme.
-
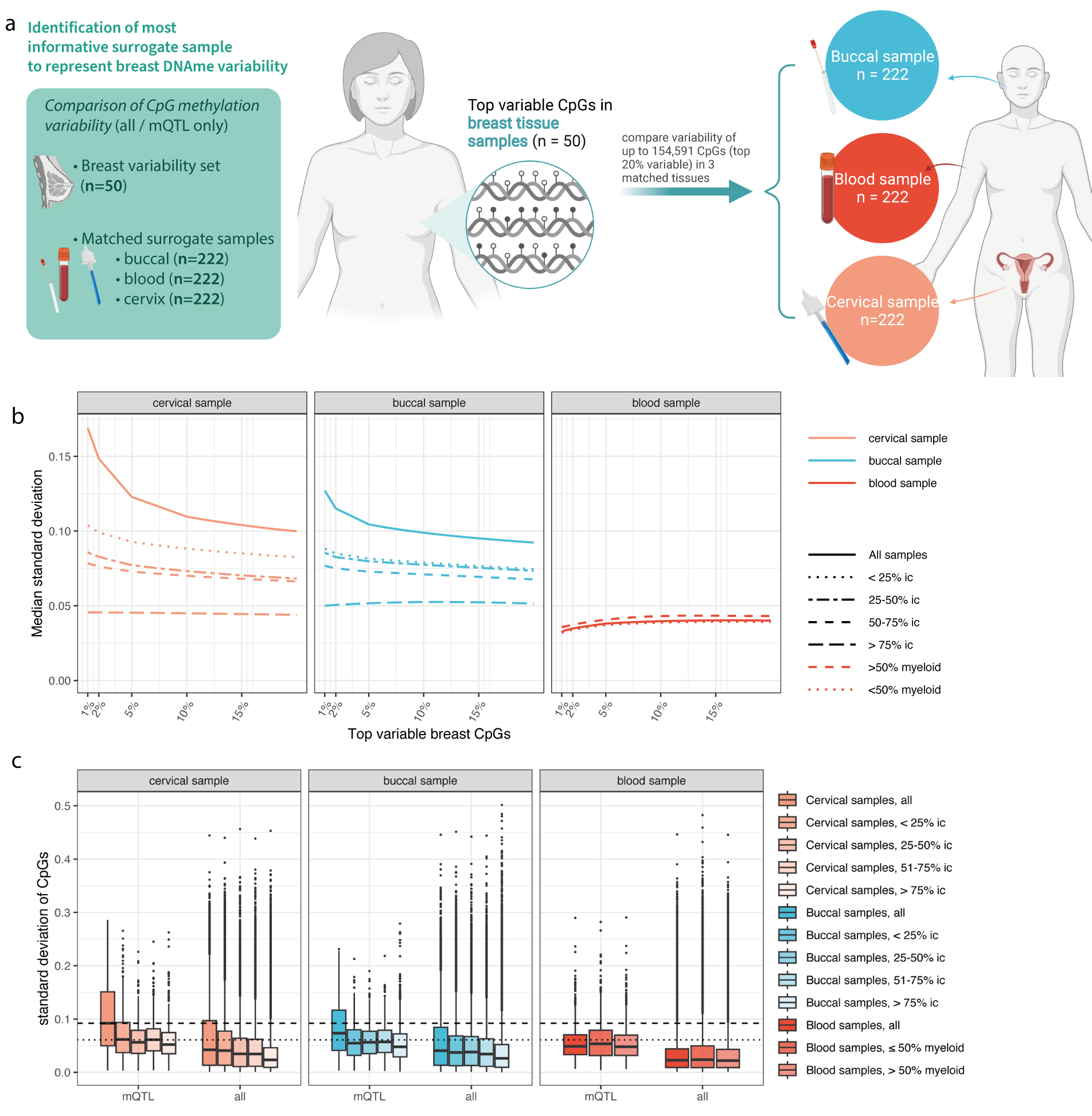 DNA methylation at quantitative trait loci (mQTLs) varies with cell type and nonheritable factors and may improve breast cancer risk assessmentChiara Herzog, Allison Jones, Iona Evans, and 8 more authorsnpj Precision Oncology, Dec 2023
DNA methylation at quantitative trait loci (mQTLs) varies with cell type and nonheritable factors and may improve breast cancer risk assessmentChiara Herzog, Allison Jones, Iona Evans, and 8 more authorsnpj Precision Oncology, Dec 2023To individualise breast cancer (BC) prevention, markers to follow a person’s changing environment and health extending beyond static genetic risk scores are required. Here, we analysed cervical and breast DNA methylation (n=1848) and single nucleotide polymorphisms (n=1442) and demonstrate that a linear combination of methylation levels at 104 BC-associated methylation quantitative trait loci (mQTL) CpGs, termed the WID™-qtBC index, can identify women with breast cancer in hormone-sensitive tissues (AUC=0.71 [95% CI: 0.65–0.77] in cervical samples). Women in the highest combined risk group (high polygenic risk score and WID™-qtBC) had a 9.6-fold increased risk for BC [95% CI: 4.7–21] compared to the low-risk group and tended to present at more advanced stages. Importantly, the WID™-qtBC is influenced by non-genetic BC risk factors, including age and body mass index, and can be modified by a preventive pharmacological intervention, indicating an interaction between genome and environment recorded at the level of the epigenome. Our findings indicate that methylation levels at mQTLs in relevant surrogate tissues could enable integration of heritable and non-heritable factors for improved disease risk stratification.
-
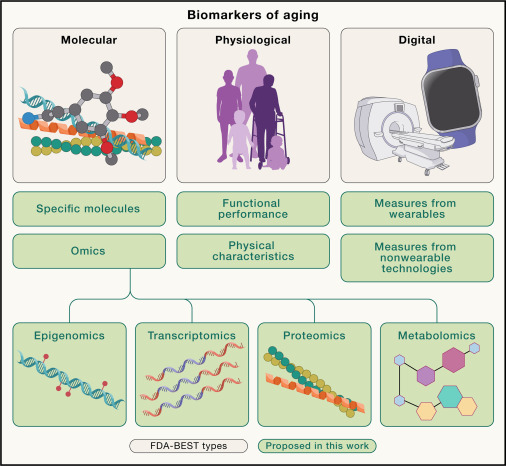 Biomarkers of aging for the identification and evaluation of longevity interventionsMahdi Moqri*, Chiara Herzog*, Jesse R. Poganik*, and 25 more authorsCell, Dec 2023
Biomarkers of aging for the identification and evaluation of longevity interventionsMahdi Moqri*, Chiara Herzog*, Jesse R. Poganik*, and 25 more authorsCell, Dec 2023With the rapid expansion of aging biology research, the identification and evaluation of longevity interventions in humans have become key goals of this field. Biomarkers of aging are critically important tools in achieving these objectives over realistic time frames. However, the current lack of standards and consensus on the properties of a reliable aging biomarker hinders their further development and validation for clinical applications. Here, we advance a framework for the terminology and characterization of biomarkers of aging, including classification and potential clinical use cases. We discuss validation steps and highlight ongoing challenges as potential areas in need of future research. This framework sets the stage for the development of valid biomarkers of aging and their ultimate utilization in clinical trials and practice.
-
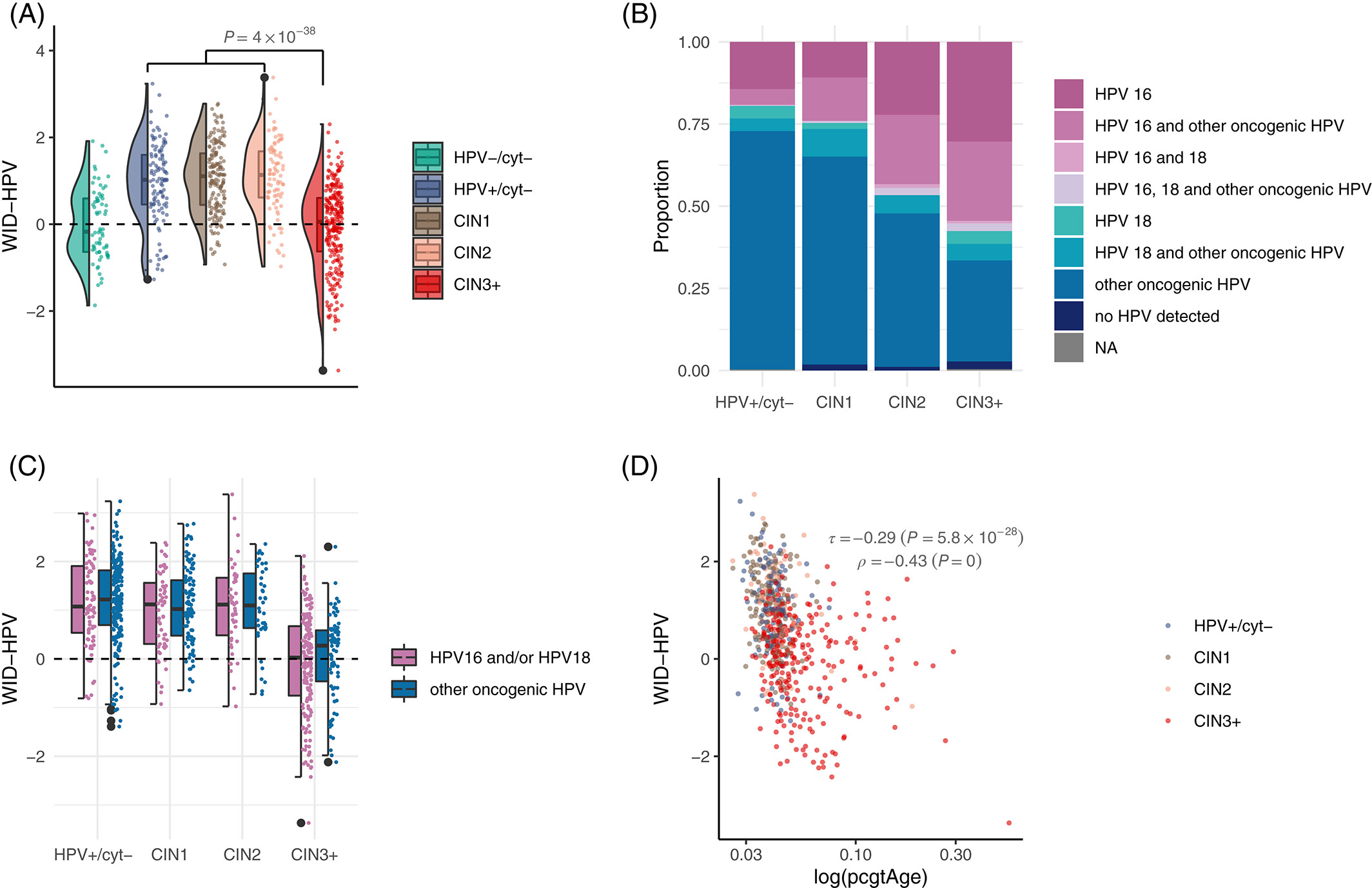 HPV‐induced host epigenetic reprogramming is lost upon progression to high‐grade cervical intraepithelial neoplasiaChiara Herzog*, Charlotte D. Vavourakis*, James E. Barrett, and 6 more authorsInternational Journal of Cancer, Dec 2023
HPV‐induced host epigenetic reprogramming is lost upon progression to high‐grade cervical intraepithelial neoplasiaChiara Herzog*, Charlotte D. Vavourakis*, James E. Barrett, and 6 more authorsInternational Journal of Cancer, Dec 2023The impact of a pathogen on host disease can only be studied in samples covering the entire spectrum of pathogenesis. Persistent oncogenic Human Papilloma Virus (HPV) infection is the most common cause for cervical cancer. Here, we investigate HPV‐induced host epigenome‐wide changes prior to development of cytological abnormalities. Using cervical sample methylation array data from disease‐free women with or without an oncogenic HPV infection, we develop the WID (Women’s cancer risk identification)‐HPV, a signature reflective of changes in the healthy host epigenome related to high‐risk HPV strains (AUC=0.78, 95% CI: 0.72‐0.85, in non‐diseased women). Looking at HPV‐associated changes across disease development, HPV‐infected women with minor cytological alterations (cervical intraepithelial neoplasia grade 1/2, CIN1/2), but surprisingly not those with precancerous changes or invasive cervical cancer (CIN3+), show an increased WID‐HPV index, indicating the WID‐HPV may reflect a successful viral clearance response absent in progression to cancer. Further investigation revealed the WID‐HPV is positively associated with apoptosis (ρ=0.48; p<0.001) and negatively associated with epigenetic replicative age (ρ=‐0.43; p<0.001). Taken together, our data suggest the WID‐HPV captures a clearance response associated with apoptosis of HPV‐infected cells. This response may be dampened or lost with increased underlying replicative age of infected cells, resulting in progression to cancer. This article is protected by copyright. All rights reserved.
-
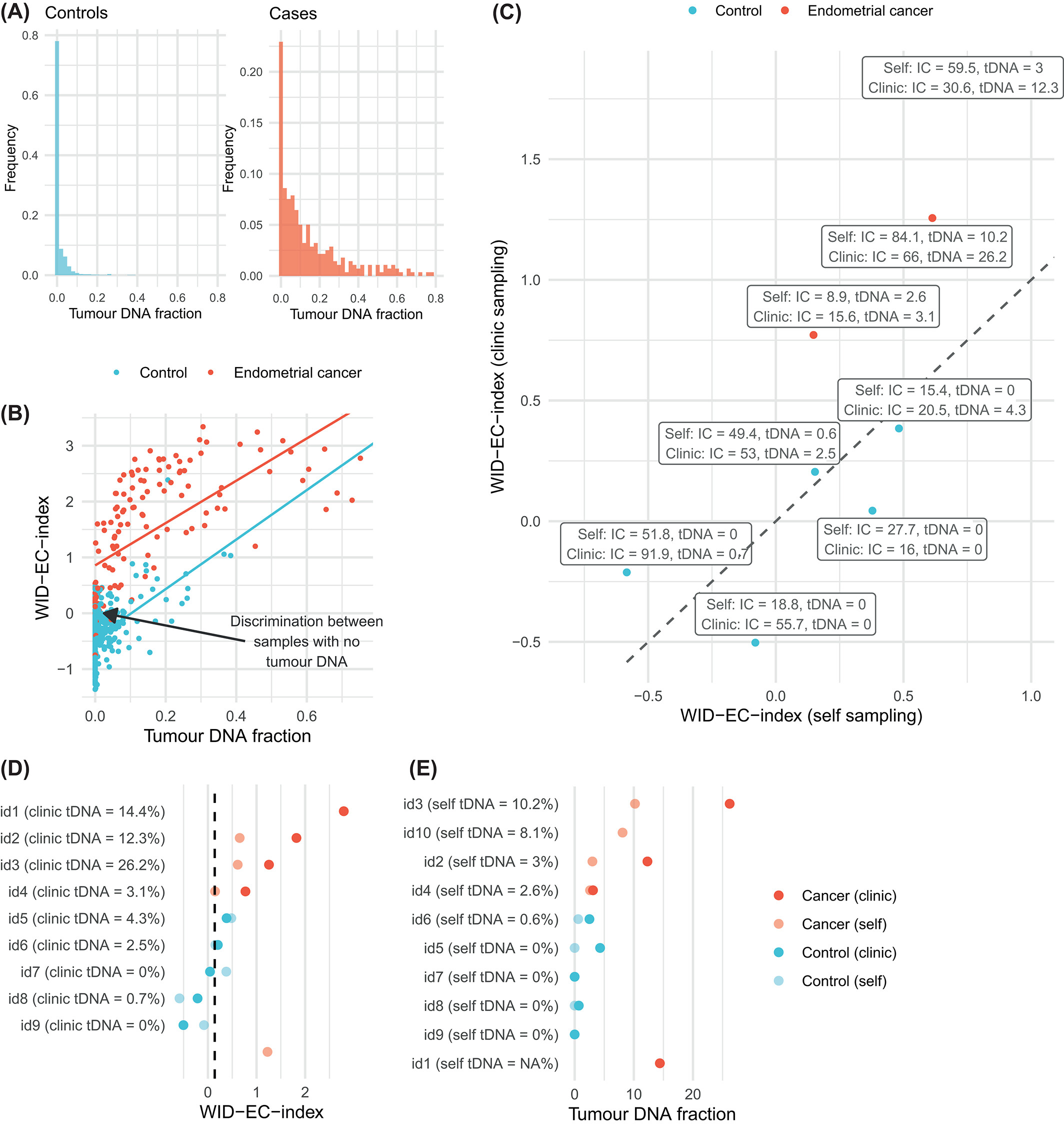 The WID‐EC test for the detection and risk prediction of endometrial cancerJames E. Barrett*, Allison Jones*, Iona Evans*, and 21 more authorsInternational Journal of Cancer, Dec 2023
The WID‐EC test for the detection and risk prediction of endometrial cancerJames E. Barrett*, Allison Jones*, Iona Evans*, and 21 more authorsInternational Journal of Cancer, Dec 2023The incidence of endometrial cancer is rising. Measures to identify women at risk and to detect endometrial cancer earlier are required to reduce the morbidity triggered by the aggressive treatment required for advanced endometrial cancer. We developed the WID‐EC (Women’s cancer risk IDentification‐Endometrial Cancer) test, which is based on DNA methylation at 500 CpG sites, in a discovery set of cervical liquid‐based cytology samples from 1086 women with and without an endometrial cancer (217 cancer cases and 869 healthy controls) with a worse prognosis (grade 3 or ≥stage IB). We validated the WID‐EC test in an independent external validation set of 64 endometrial cancer cases and 225 controls. We further validated the test in 150 healthy women (prospective set) who provided a cervical sample as part of the routine Swedish cervical screening programme, 54 of whom developed endometrial cancer within 3 years of sample collection. The WID‐EC test identified women with endometrial cancer with a receiver operator characteristic area under the curve (AUC) of 0.92 (95% CI: 0.88‐0.97) in the external set and of 0.82 (95% CI: 0.74‐0.89) in the prospective validation set. Using an optimal cutoff, cancer cases were detected with a sensitivity of 86% and a specificity of 90% in the external validation set, and a sensitivity and specificity of 52% and 98% respectively in the prospective validation set. The WID‐EC test can identify women with or at risk of endometrial cancer. The endometrial cancer incidence is rising, and imaging‐based screening is often not suitably accurate for endometrial cancers. Here, the authors show that a new DNA methylation signature (Women’s cancer risk IDentification‐Endometrial Cancer, or WID‐EC) can both detect the presence and predict the risk of endometrial cancer. Together with three other recently‐reported methylation indices that detect breast, ovarian, and cervical cancers (WID‐BC, WID‐OC, and WID‐CIN), WID‐EC completes a list of signatures that can detect/predict all four women‐specific cancers using a single cervical sample and the same technology platform for methylation array‐based tests.
2022
-
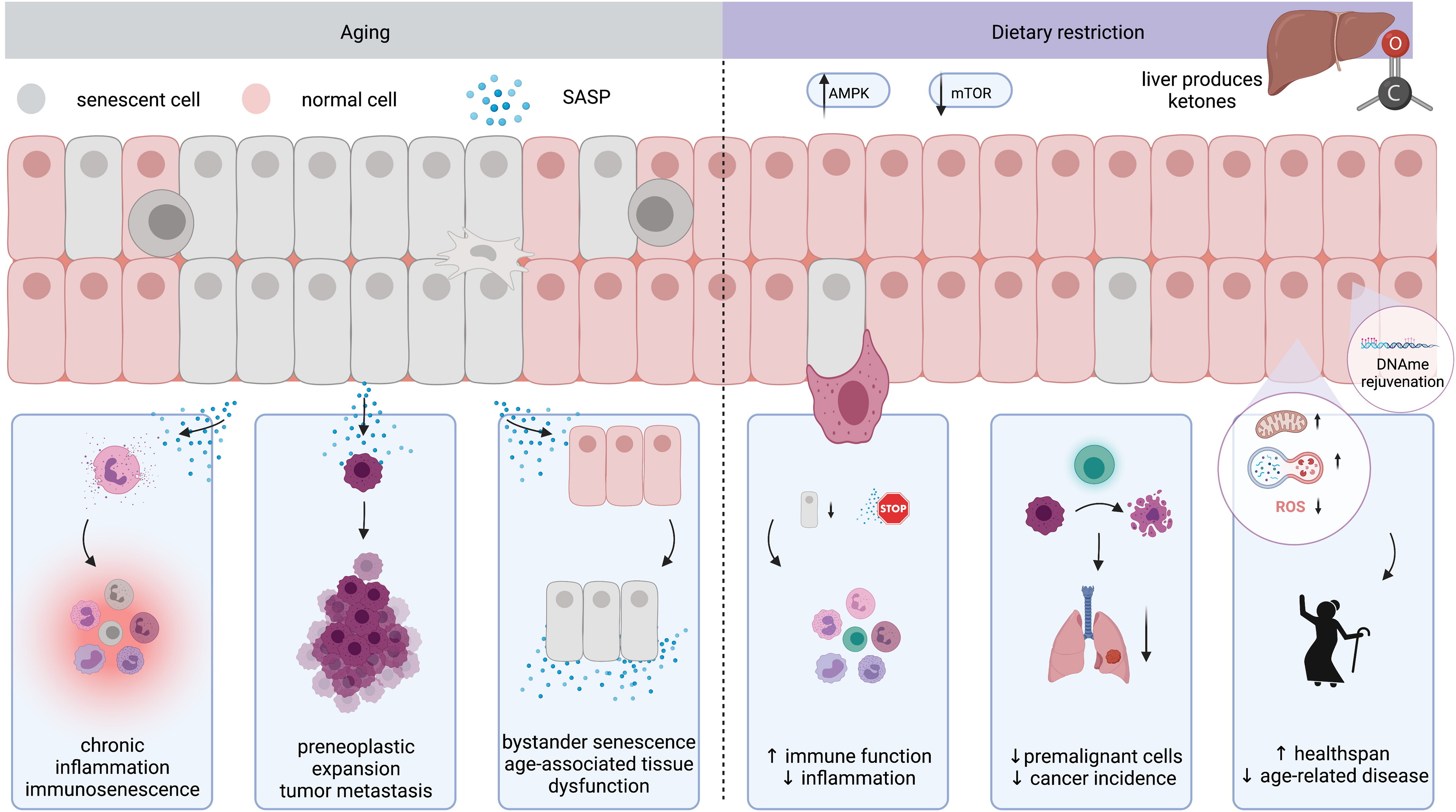 Dietary restriction in senolysis and prevention and treatment of diseaseSepideh Aminzadeh-Gohari, Barbara Kofler, and Chiara HerzogCritical Reviews in Food Science and Nutrition, Dec 2022
Dietary restriction in senolysis and prevention and treatment of diseaseSepideh Aminzadeh-Gohari, Barbara Kofler, and Chiara HerzogCritical Reviews in Food Science and Nutrition, Dec 2022Aging represents a key risk factor for a plethora of diseases. Targeting detrimental processes which occur during aging, especially before onset of age-related disease, could provide drastic improvements in healthspan. There is increasing evidence that dietary restriction (DR), including caloric restriction, fasting, or fasting-mimicking diets, extend both lifespan and healthspan. This has sparked interest in the use of dietary regimens as a non-pharmacological means to slow aging and prevent disease. Here, we review the current evidence on the molecular mechanisms underlying DR-induced health improvements, including removal of senescent cells, metabolic reprogramming, and epigenetic rejuvenation.
-
 A Simple Cervicovaginal Epigenetic Test for Screening and Rapid Triage of Women With Suspected Endometrial Cancer: Validation in Several Cohort and Case/Control SetsChiara Herzog*, Fátima Marín*, Allison Jones, and 28 more authorsJournal of Clinical Oncology, Dec 2022
A Simple Cervicovaginal Epigenetic Test for Screening and Rapid Triage of Women With Suspected Endometrial Cancer: Validation in Several Cohort and Case/Control SetsChiara Herzog*, Fátima Marín*, Allison Jones, and 28 more authorsJournal of Clinical Oncology, Dec 2022PURPOSE: Endometrial cancer (EC) incidence has been rising over the past 10 years. Delays in diagnosis reduce survival and necessitate more aggressive treatment. We aimed to develop and validate a simple, noninvasive, and reliable triage test for EC to reduce the number of invasive diagnostic procedures and improve patient survival.
METHODS: We developed a test to screen and triage women with suspected EC using 726 cervical smear samples from women with and without EC, and validated the test in 562 cervicovaginal samples using three different collection methods (cervical smear: n = 248; vaginal swab: n = 63; and self-collection: n = 251) and four different settings (case/control: n = 388; cohort of women presenting with postmenopausal bleeding: n = 63; a cohort of high-risk women with Lynch syndrome: n = 25; and a nested case/control setting from a screening cohort and samples taken up to 3 years before EC diagnosis: n = 86).
RESULTS: We describe the Women’s cancer risk IDentification – quantitative polymerase chain reaction test for Endometrial Cancer (WID-qEC), a three-marker test that evaluates DNA methylation in gene regions of GYPC and ZSCAN12. In cervical, self-collected, and vaginal swab samples derived from symptomatic patients, it detected EC with sensitivities of 97.2% (95% CI, 90.2 to 99.7), 90.1% (83.6 to 94.6), and 100% (63.1 to 100), respectively, and specificities of 75.8% (63.6 to 85.5), 86.7% (79.3 to 92.2), and 89.1% (77.8 to 95.9), respectively. The WID-qEC identified 90.9% (95% CI, 70.8 to 98.9) of EC cases in samples predating diagnosis up to 1 year. Test performance was similar across menopausal status, age, stage, grade, ethnicity, and histology.
CONCLUSION: The WID-qEC is a noninvasive reliable test for triage of women with symptoms suggestive of ECs. Because of the potential for self-collection, it could improve early diagnosis and reduce the reliance for in-person visits. -
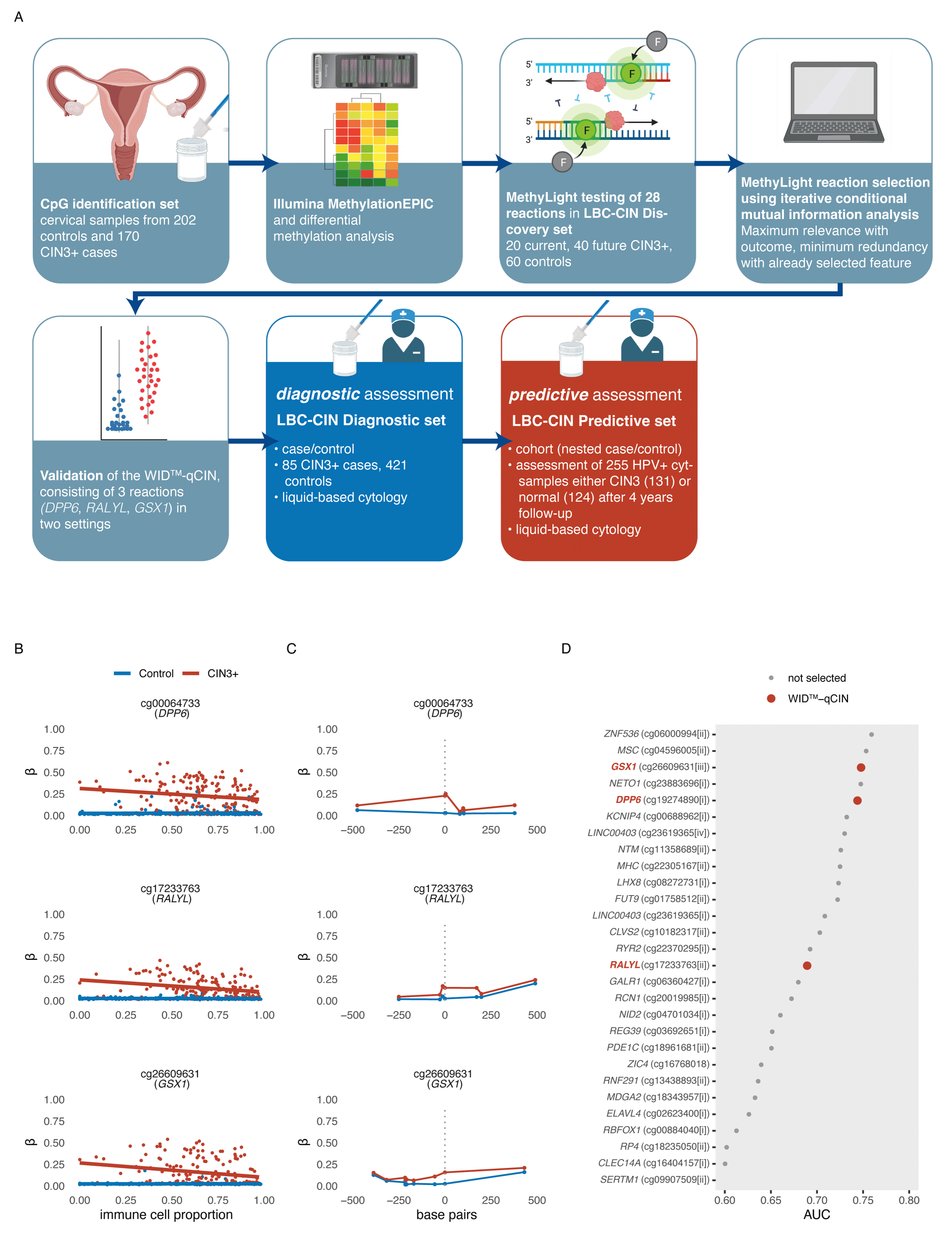 DNA methylation-based detection and prediction of cervical intraepithelial neoplasia grade 3 and invasive cervical cancer with the WID™-qCIN testChiara Herzog*, Karin Sundström*, Allison Jones, and 14 more authorsClinical Epigenetics, Dec 2022
DNA methylation-based detection and prediction of cervical intraepithelial neoplasia grade 3 and invasive cervical cancer with the WID™-qCIN testChiara Herzog*, Karin Sundström*, Allison Jones, and 14 more authorsClinical Epigenetics, Dec 2022Cervical screening using primary human papilloma virus (HPV) testing and cytology is being implemented in several countries. Cytology as triage for colposcopy referral suffers from several shortcomings. HPV testing overcomes some of these but lacks specificity in women under 30. Here, we aimed to develop and validate an automatable triage test that is highly sensitive and specific independently of age and sample heterogeneity, and predicts progression to CIN3+ in HPV+ patients. The WID™-qCIN, assessing three regions in human genes DPP6, RALYL, and GSX1, was validated in both a diagnostic (case–control) and predictive setting (nested case–control), in a total of 761 samples. Using a predefined threshold, the sensitivity of the WID™-qCIN test was 100% and 78% to detect invasive cancer and CIN3, respectively. Sensitivity to detect CIN3+ was 65% and 83% for women < and ≥ 30 years of age. The specificity was 90%. Importantly, the WID™-qCIN test identified 52% of ≥ 30-year-old women with a cytology negative (cyt−) index sample who were diagnosed with CIN3 1–4 years after sample donation. We identified suitable DNAme regions in an epigenome-wide discovery using HPV+ controls and CIN3+ cases and established the WID™-qCIN, a PCR-based DNAme test. The WID™-qCIN test has a high sensitivity and specificity that may outperform conventional cervical triage tests and can in an objective, cheap, and scalable fashion identify most women with and at risk of (pre-)invasive cervical cancer. However, evaluation was limited to case–control settings and future studies will assess performance and generalisability in a randomised controlled trial.
-
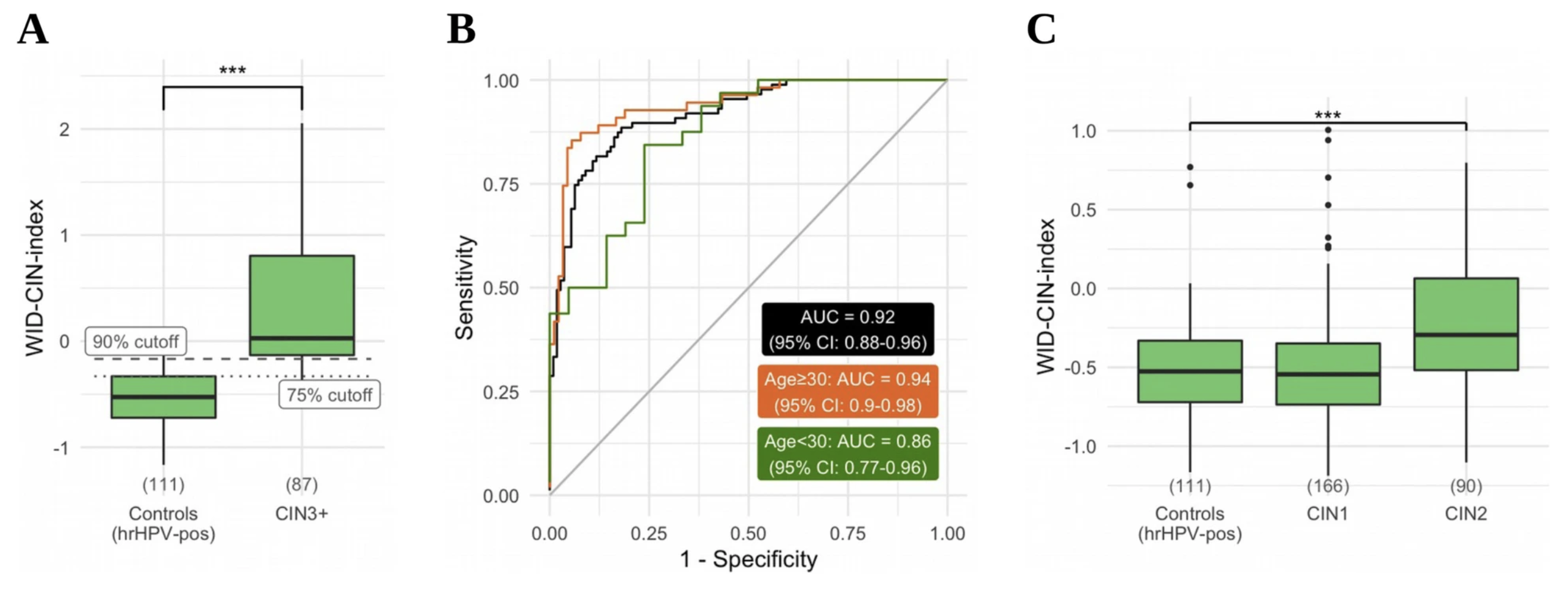 The WID-CIN test identifies women with, and at risk of, cervical intraepithelial neoplasia grade 3 and invasive cervical cancerJames E. Barrett, Karin Sundström, Allison Jones, and 5 more authorsGenome Medicine, Dec 2022
The WID-CIN test identifies women with, and at risk of, cervical intraepithelial neoplasia grade 3 and invasive cervical cancerJames E. Barrett, Karin Sundström, Allison Jones, and 5 more authorsGenome Medicine, Dec 2022Cervical screening is transitioning from primary cytology to primary human papillomavirus (HPV) testing. HPV testing is highly sensitive but there is currently no high-specificity triage method for colposcopy referral to detect cervical intraepithelial neoplasia grade 3 or above (CIN3+) in women positive for high-risk (hr) HPV subtypes. An objective, automatable test that could accurately perform triage, independently of sample heterogeneity and age, is urgently required. We analyzed DNA methylation at 850,000 CpG sites across the genome in a total of 1254 cervical liquid-based cytology (LBC) samples from cases of screen-detected histologically verified CIN1-3+ (98% hrHPV-positive) and population-based control women free from any cervical disease (100% hrHPV-positive). Samples were provided by a state-of-the-art population-based cohort biobank and consisted of (i) a discovery set of 170 CIN3+ cases and 202 hrHPV-positive/cytology-negative controls; (ii) a diagnostic validation set of 87 CIN3+, 90 CIN2, 166 CIN1, and 111 hrHPV-positive/cytology-negative controls; and (iii) a predictive validation set of 428 cytology-negative samples (418 hrHPV-positive) of which 210 were diagnosed with CIN3+ in the upcoming 1–4 years and 218 remained disease-free. We developed the WID-CIN (Women’s cancer risk IDentification-Cervical Intraepithelial Neoplasia) test, a DNA methylation signature consisting of 5000 CpG sites. The receiver operating characteristic area under the curve (AUC) in the independent diagnostic validation set was 0.92 (95% CI 0.88–0.96). At 75% specificity (≤CIN1), the overall sensitivity to detect CIN3+ is 89.7% (83.3–96.1) in all and 92.7% (85.9–99.6) and 65.6% (49.2–82.1) in women aged ≥30 and <30. In hrHPV-positive/cytology-negative samples in the predictive validation set, the WID-CIN detected 54.8% (48.0–61.5) cases developing 1–4 years after sample donation in all ages or 56.9% (47.6–66.2) and 53.5% (43.7–63.2) in ≥30 and <30-year-old women, at a specificity of 75%. The WID-CIN test identifies the vast majority of hrHPV-positive women with current CIN3+ lesions. In the absence of cytologic abnormalities, a positive WID-CIN test result is likely to indicate a significantly increased risk of developing CIN3+ in the near future.
-
 The WID‐qEC test: Performance in a hospital‐based cohort and feasibility to detect endometrial and cervical cancersLena Schreiberhuber, Chiara Herzog, Charlotte D. Vavourakis, and 13 more authorsInternational Journal of Cancer, Dec 2022
The WID‐qEC test: Performance in a hospital‐based cohort and feasibility to detect endometrial and cervical cancersLena Schreiberhuber, Chiara Herzog, Charlotte D. Vavourakis, and 13 more authorsInternational Journal of Cancer, Dec 2022The majority of endometrial and cervical cancers present with abnormal vaginal bleeding but only a small proportion of women suffering from vaginal bleeding actually have such a cancer. A simple, operator‐independent and accurate test to correctly identify women presenting with abnormal bleeding as a consequence of endometrial or cervical cancer is urgently required. We have recently developed and validated the WID‐qEC test, which assesses DNA methylation of ZSCAN12 and GYPC via real‐time PCR, to triage women with symptoms suggestive of endometrial cancer using ThinPrep‐based liquid cytology samples. Here, we investigated whether the WID‐qEC test can additionally identify women with cervical cancer. Moreover, we evaluate the test’s applicability in a SurePath‐based hospital‐cohort by comparing its ability to detect endometrial and cervical cancer to cytology. In a set of 23 cervical cancer cases and 28 matched controls the receiver operating characteristic (ROC) area under the curve (AUC) is 0.99 (95% confidence interval [CI]: 0.97‐1.00) with a sensitivity and specificity of 100% and 92.9%, respectively. Amongst the hospital‐cohort (n = 330), the ROC AUC is 0.99 (95% CI: 0.98‐1) with a sensitivity and specificity of 100% and 82.5% for the WID‐qEC test, respectively, and 33.3% and 96.9% for cytology (considering PAP IV/V as positive). Our data suggest that the WID‐qEC test detects both endometrial and cervical cancer with high accuracy. While abnormal vaginal bleeding is a presenting symptom of endometrial and cervical cancers, only a small proportion of women who present with vaginal bleeding have endometrial or cervical cancer. Currently, the tests used to triage women with abnormal bleeding, such as ultrasound or cytology, are subjective and have modest accuracy. Here, the authors demonstrate that a real‐time PCR‐based test, which assesses DNA methylation at three gene regions using a cervical or vaginal sample, is able to identify 100% of women who have cervical or endometrial cancer with a high specificity (>80%), irrespective of the sample collection system.
-
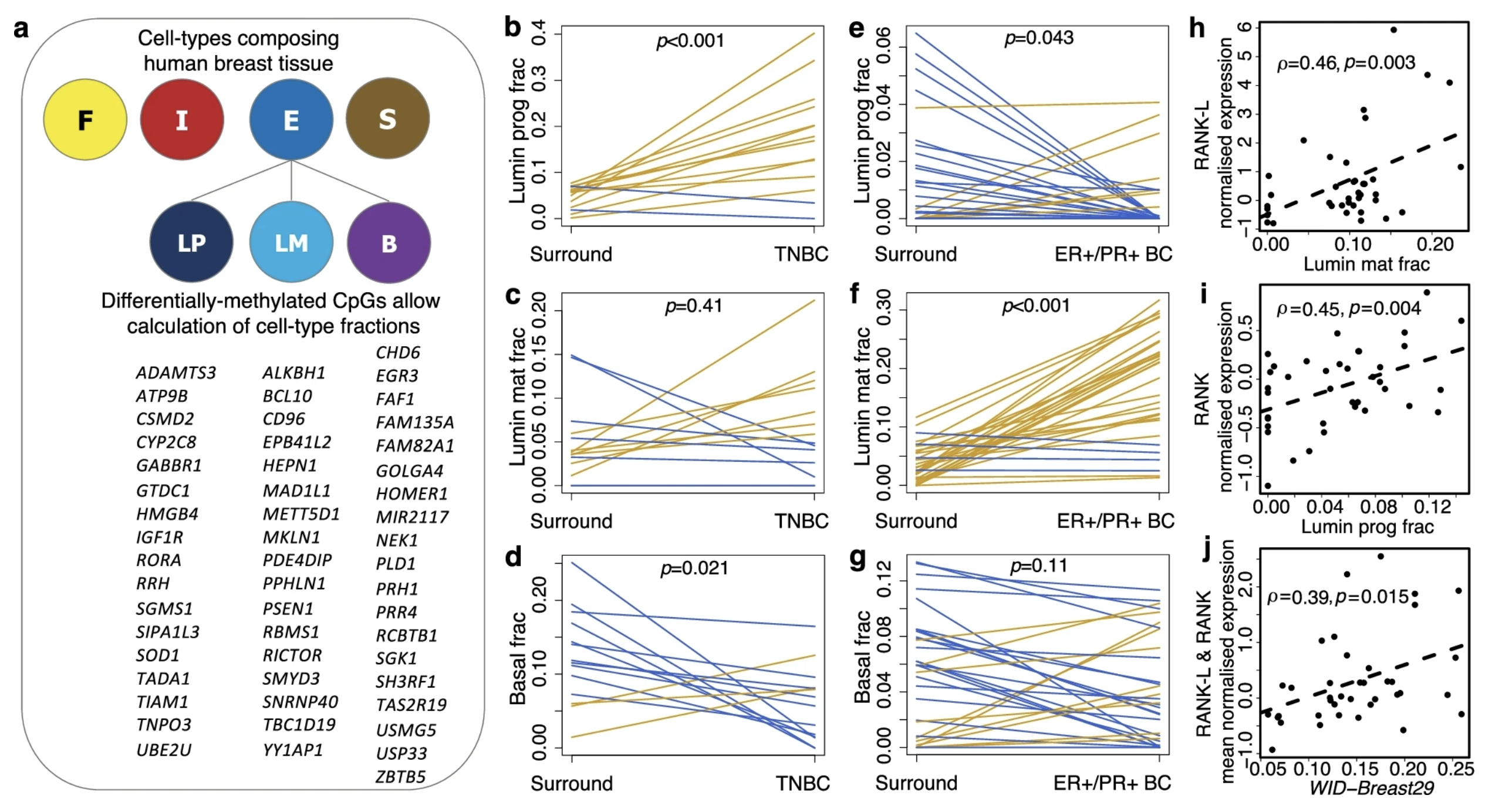 Antiprogestins reduce epigenetic field cancerization in breast tissue of young healthy womenThomas E. Bartlett, Iona Evans, Allison Jones, and 20 more authorsGenome Medicine, Dec 2022
Antiprogestins reduce epigenetic field cancerization in breast tissue of young healthy womenThomas E. Bartlett, Iona Evans, Allison Jones, and 20 more authorsGenome Medicine, Dec 2022Breast cancer is a leading cause of death in premenopausal women. Progesterone drives expansion of luminal progenitor cells, leading to the development of poor-prognostic breast cancers. However, it is not known if antagonising progesterone can prevent breast cancers in humans. We suggest that targeting progesterone signalling could be a means of reducing features which are known to promote breast cancer formation. In healthy premenopausal women with and without a BRCA mutation we studied (i) estrogen and progesterone levels in saliva over an entire menstrual cycle (n = 20); (ii) cancer-free normal breast-tissue from a control population who had no family or personal history of breast cancer and equivalently from BRCA1/2 mutation carriers (n = 28); triple negative breast cancer (TNBC) biopsies and healthy breast tissue taken from sites surrounding the TNBC in the same individuals (n = 14); and biopsies of ER+ve/PR+ve stage T1–T2 cancers and healthy breast tissue taken from sites surrounding the cancer in the same individuals (n = 31); and (iii) DNA methylation and DNA mutations in normal breast tissue (before and after treatment) from clinical trials that assessed the potential preventative effects of vitamins and antiprogestins (mifepristone and ulipristal acetate; n = 44). Daily levels of progesterone were higher throughout the menstrual cycle of BRCA1/2 mutation carriers, raising the prospect of targeting progesterone signalling as a means of cancer risk reduction in this population. Furthermore, breast field cancerization DNA methylation signatures reflective of (i) the mitotic age of normal breast epithelium and (ii) the proportion of luminal progenitor cells were increased in breast cancers, indicating that luminal progenitor cells with elevated replicative age are more prone to malignant transformation. The progesterone receptor antagonist mifepristone reduced both the mitotic age and the proportion of luminal progenitor cells in normal breast tissue of all control women and in 64% of BRCA1/2 mutation carriers. These findings were validated by an alternate progesterone receptor antagonist, ulipristal acetate, which yielded similar results. Importantly, mifepristone reduced both the TP53 mutation frequency as well as the number of TP53 mutations in mitotic-age-responders. These data support the potential usage of antiprogestins for primary prevention of poor-prognostic breast cancers. Clinical trial 1 Mifepristone treatment prior to insertion of a levonorgestrel releasing intrauterine system for improved bleeding control – a randomized controlled trial, clinicaltrialsregister.eu, 2009-009014-40; registered on 20 July 2009. Clinical trial 2 The effect of a progesterone receptor modulator on breast tissue in women with BRCA1 and 2 mutations, clinicaltrials.gov, NCT01898312; registered on 07 May 2013. Clinical trial 3 A pilot prevention study of the effects of the anti- progestin Ulipristal Acetate (UA) on surrogate markers of breast cancer risk, clinicaltrialsregister.eu, 2015-001587-19; registered on 15 July 2015.
-
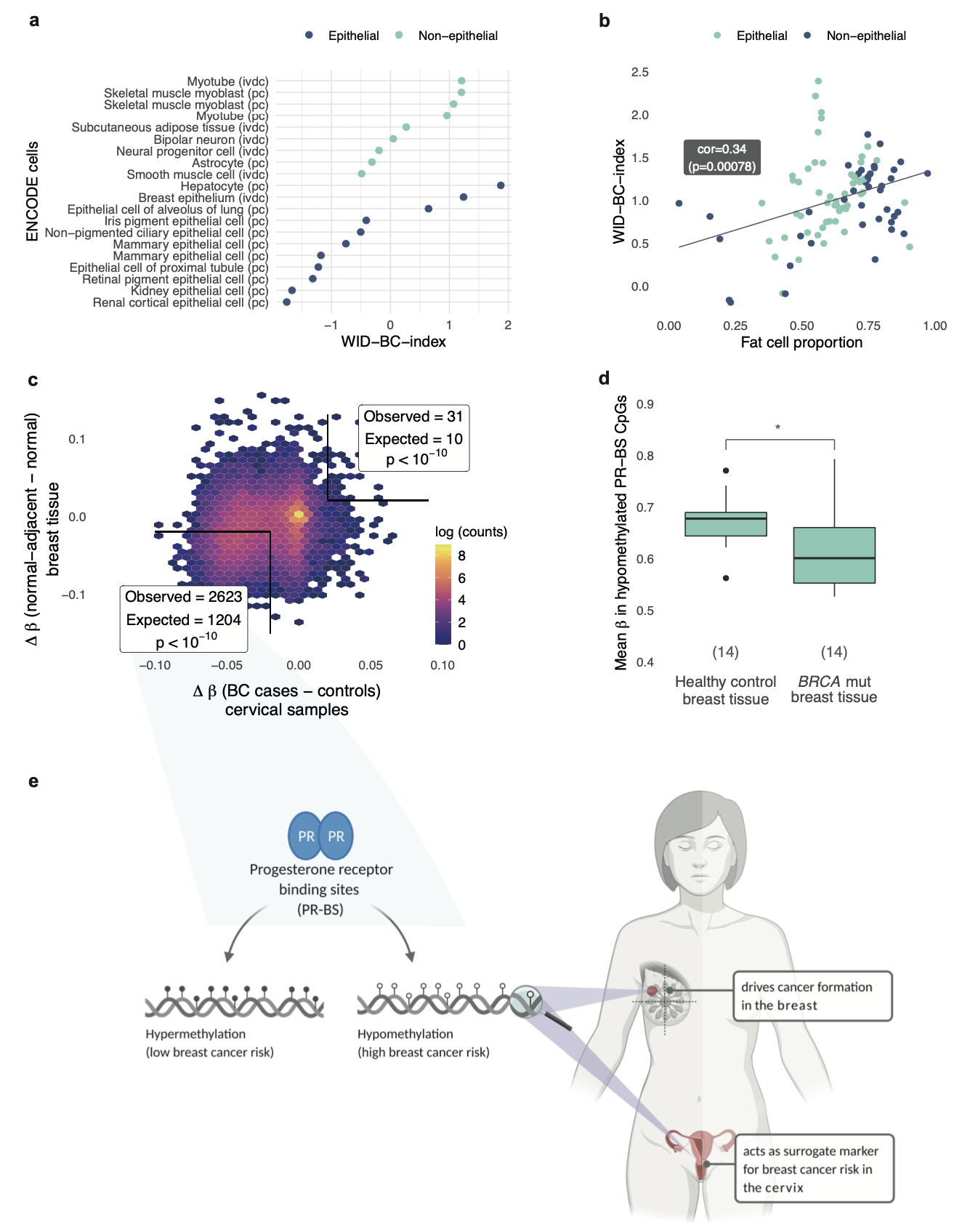 The WID-BC-index identifies women with primary poor prognostic breast cancer based on DNA methylation in cervical samplesJames E. Barrett*, Chiara Herzog*, Allison Jones, and 22 more authorsNature Communications, Dec 2022
The WID-BC-index identifies women with primary poor prognostic breast cancer based on DNA methylation in cervical samplesJames E. Barrett*, Chiara Herzog*, Allison Jones, and 22 more authorsNature Communications, Dec 2022Genetic and non-genetic factors contribute to breast cancer development. An epigenome-based signature capturing these components in easily accessible samples could identify women at risk. Here, we analyse the DNA methylome in 2,818 cervical, 357 and 227 matched buccal and blood samples respectively, and 42 breast tissue samples from women with and without breast cancer. Utilising cervical liquid-based cytology samples, we develop the DNA methylation-based Women’s risk IDentification for Breast Cancer index (WID-BC-index) that identifies women with breast cancer with an AUROC (Area Under the Receiver Operator Characteristic) of 0.84 (95% CI: 0.80–0.88) and 0.81 (95% CI: 0.76–0.86) in internal and external validation sets, respectively. CpGs at progesterone receptor binding sites hypomethylated in normal breast tissue of women with breast cancer or in BRCA mutation carriers are also hypomethylated in cervical samples of women with poor prognostic breast cancer. Our data indicate that a systemic epigenetic programming defect is highly prevalent in women who develop breast cancer. Further studies validating the WID-BC-index may enable clinical implementation for monitoring breast cancer risk. Breast cancer is most commonly diagnosed via a needle biopsy. In this study, the authors show that cervical samples from women with breast cancer have a methylation signature different to that of healthy controls.
-
 The DNA methylome of cervical cells can predict the presence of ovarian cancerJames E. Barrett, Allison Jones, Iona Evans, and 10 more authorsNature Communications, Dec 2022
The DNA methylome of cervical cells can predict the presence of ovarian cancerJames E. Barrett, Allison Jones, Iona Evans, and 10 more authorsNature Communications, Dec 2022The vast majority of epithelial ovarian cancer arises from tissues that are embryologically derived from the Müllerian Duct. Here, we demonstrate that a DNA methylation signature in easy-to-access Müllerian Duct-derived cervical cells from women with and without ovarian cancer (i.e. referred to as the Women’s risk IDentification for Ovarian Cancer index or WID-OC-index) is capable of identifying women with an ovarian cancer in the absence of tumour DNA with an AUC of 0.76 and women with an endometrial cancer with an AUC of 0.81. This and the observation that the cervical cell WID-OC-index mimics the epigenetic program of those cells at risk of becoming cancerous in BRCA1/2 germline mutation carriers (i.e. mammary epithelium, fallopian tube fimbriae, prostate) further suggest that the epigenetic misprogramming of cervical cells is an indicator for cancer predisposition. This concept has the potential to advance the field of risk-stratified cancer screening and prevention. Most ovarian cancers originate from cells originally derived from Müllerian Duct cells. Here, the authors show that the methylation profile of Müllerian Duct cells isolated from cervical samples can predict whether a woman has cervical cancer.
-
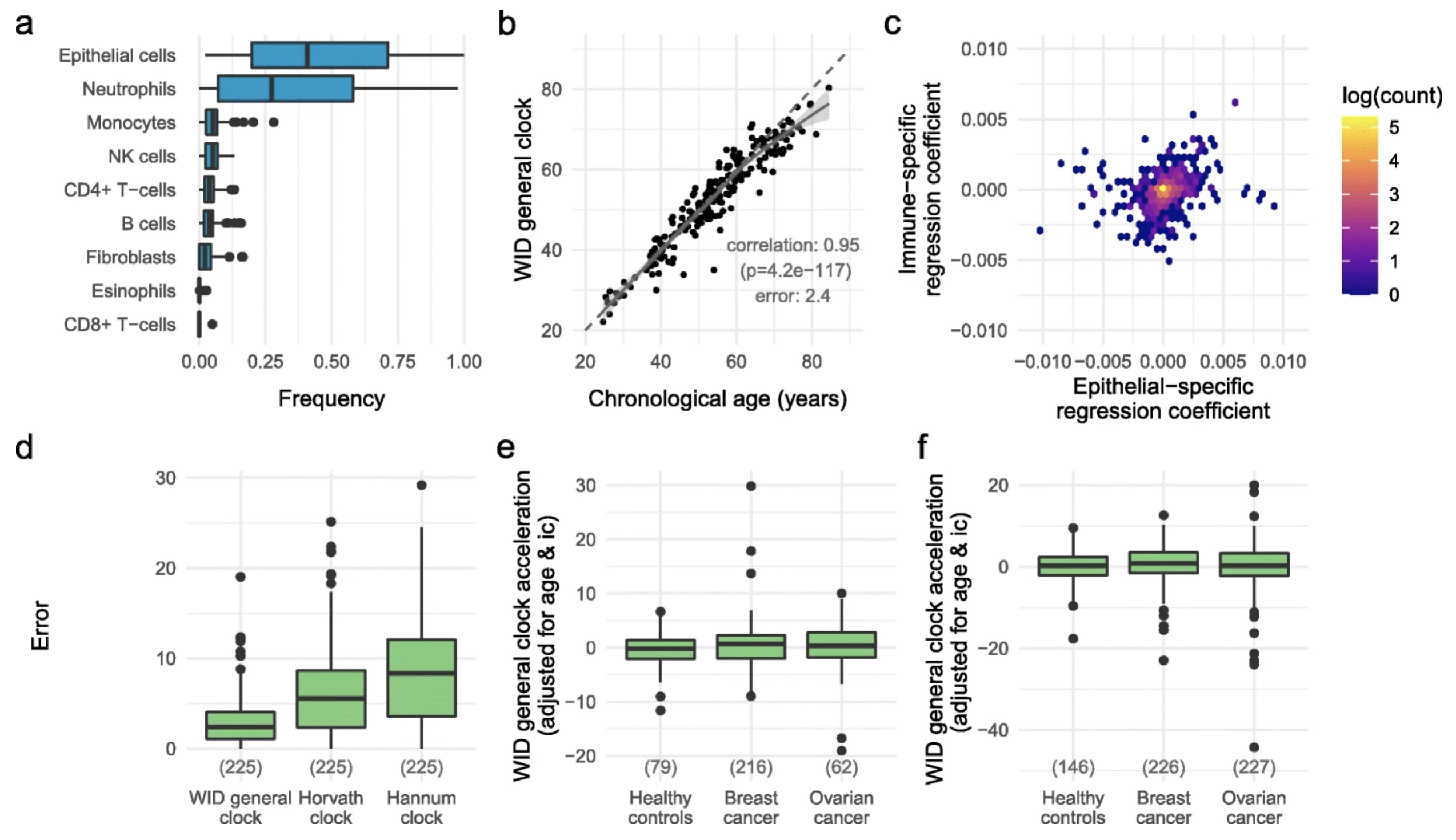 Susceptibility to hormone-mediated cancer is reflected by different tick rates of the epithelial and general epigenetic clockJames E. Barrett*, Chiara Herzog*, Yoo-Na Kim*, and 12 more authorsGenome Biology, Dec 2022
Susceptibility to hormone-mediated cancer is reflected by different tick rates of the epithelial and general epigenetic clockJames E. Barrett*, Chiara Herzog*, Yoo-Na Kim*, and 12 more authorsGenome Biology, Dec 2022A variety of epigenetic clocks utilizing DNA methylation changes have been developed; these clocks are either tissue-independent or designed to predict chronological age based on blood or saliva samples. Whether discordant tick rates between tissue-specific and general epigenetic clocks play a role in health and disease has not yet been explored. Here we analyze 1941 cervical cytology samples, which contain a mixture of hormone-sensitive cervical epithelial cells and immune cells, and develop the WID general clock (Women’s IDentification of risk), an epigenetic clock that is shared by epithelial and immune cells and optimized for cervical samples. We then develop the WID epithelial clock and WID immune clock, which define epithelial- and immune-specific clocks, respectively. We find that the WID-relative-epithelial-age (WID-REA), defined as the difference between the epithelial and general clocks, is significantly reduced in cervical samples from pre-menopausal women with breast cancer (OR 2.7, 95% CI 1.28-5.72). We find the same effect in normal breast tissue samples from pre-menopausal women at high risk of breast cancer and show that potential risk reducing anti-progesterone drugs can reverse this. In post-menopausal women, this directionality is reversed. Hormone replacement therapy consistently leads to a significantly lower WID-REA in cancer-free women, but not in post-menopausal women with breast or ovarian cancer. Our findings imply that there are multiple epigenetic clocks, many of which are tissue-specific, and that the differential tick rate between these clocks may be an informative surrogate measure of disease risk.